Structural and dynamical description of the enzymatic reaction of a phosphohexomutase
Abstract
Enzymes are known to adopt various conformations at different points along their catalytic cycles. Here, we present a comprehensive analysis of 15 isomorphous, high resolution crystal structures of the enzyme phosphoglucomutase from the bacterium Xanthomonas citri. The protein was captured in distinct states critical to function, including enzyme-substrate, enzyme-product, and enzyme-intermediate complexes. Key residues in ligand recognition and regions undergoing conformational change are identified and correlated with the various steps of the catalytic reaction. In addition, we use principal component analysis to examine various subsets of these structures with two goals: (1) identifying sites of conformational heterogeneity through a comparison of room temperature and cryogenic structures of the apo-enzyme and (2) a priori clustering of the enzyme-ligand complexes into functionally related groups, showing sensitivity of this method to structural features difficult to detect by traditional methods. This study captures, in a single system, the structural basis of diverse substrate recognition, the subtle impact of covalent modification, and the role of ligand-induced conformational change in this representative enzyme of the α-D-phosphohexomutase superfamily.
- Authors:
-
- Univ. of Missouri, Columbia, MO (United States)
- Dalhousie Univ., Halifax, NS (Canada)
- Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States). Advanced Light Source (ALS)
- Publication Date:
- Research Org.:
- Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC), Basic Energy Sciences (BES); National Institutes of Health (NIH); National Science Foundation (NSF)
- OSTI Identifier:
- 1619116
- Alternate Identifier(s):
- OSTI ID: 1504500
- Grant/Contract Number:
- AC02-05CH11231; T32 GM008396-26; MCB-0918389; P30 GM124169-01
- Resource Type:
- Accepted Manuscript
- Journal Name:
- Structural Dynamics
- Additional Journal Information:
- Journal Volume: 6; Journal Issue: 2; Journal ID: ISSN 2329-7778
- Publisher:
- American Crystallographic Association/AIP
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 37 INORGANIC, ORGANIC, PHYSICAL, AND ANALYTICAL CHEMISTRY
Citation Formats
Stiers, Kyle M., Graham, Abigail C., Zhu, Jian-She, Jakeman, David L., Nix, Jay C., and Beamer, Lesa J. Structural and dynamical description of the enzymatic reaction of a phosphohexomutase. United States: N. p., 2019.
Web. doi:10.1063/1.5092803.
Stiers, Kyle M., Graham, Abigail C., Zhu, Jian-She, Jakeman, David L., Nix, Jay C., & Beamer, Lesa J. Structural and dynamical description of the enzymatic reaction of a phosphohexomutase. United States. https://doi.org/10.1063/1.5092803
Stiers, Kyle M., Graham, Abigail C., Zhu, Jian-She, Jakeman, David L., Nix, Jay C., and Beamer, Lesa J. Mon .
"Structural and dynamical description of the enzymatic reaction of a phosphohexomutase". United States. https://doi.org/10.1063/1.5092803. https://www.osti.gov/servlets/purl/1619116.
@article{osti_1619116,
title = {Structural and dynamical description of the enzymatic reaction of a phosphohexomutase},
author = {Stiers, Kyle M. and Graham, Abigail C. and Zhu, Jian-She and Jakeman, David L. and Nix, Jay C. and Beamer, Lesa J.},
abstractNote = {Enzymes are known to adopt various conformations at different points along their catalytic cycles. Here, we present a comprehensive analysis of 15 isomorphous, high resolution crystal structures of the enzyme phosphoglucomutase from the bacterium Xanthomonas citri. The protein was captured in distinct states critical to function, including enzyme-substrate, enzyme-product, and enzyme-intermediate complexes. Key residues in ligand recognition and regions undergoing conformational change are identified and correlated with the various steps of the catalytic reaction. In addition, we use principal component analysis to examine various subsets of these structures with two goals: (1) identifying sites of conformational heterogeneity through a comparison of room temperature and cryogenic structures of the apo-enzyme and (2) a priori clustering of the enzyme-ligand complexes into functionally related groups, showing sensitivity of this method to structural features difficult to detect by traditional methods. This study captures, in a single system, the structural basis of diverse substrate recognition, the subtle impact of covalent modification, and the role of ligand-induced conformational change in this representative enzyme of the α-D-phosphohexomutase superfamily.},
doi = {10.1063/1.5092803},
journal = {Structural Dynamics},
number = 2,
volume = 6,
place = {United States},
year = {Mon Apr 01 00:00:00 EDT 2019},
month = {Mon Apr 01 00:00:00 EDT 2019}
}
Web of Science
Figures / Tables:
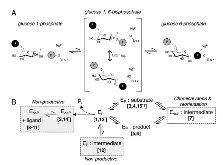 FIG. 1: Overview of the mechanism and structure of XcPGM. (a) A schematic of the catalytic reaction, showing the reversible conversion of glucose 1-phosphate to glucose 6-phosphate. Glucose 1,6-bisphosphate undergoes a 180° reorientation in between the two phosphoryl transfer steps of the reaction. (b) An outline of the catalytic cyclemore »
FIG. 1: Overview of the mechanism and structure of XcPGM. (a) A schematic of the catalytic reaction, showing the reversible conversion of glucose 1-phosphate to glucose 6-phosphate. Glucose 1,6-bisphosphate undergoes a 180° reorientation in between the two phosphoryl transfer steps of the reaction. (b) An outline of the catalytic cyclemore »
Works referenced in this record:
Identification of an essential active-site residue in the α- d -phosphohexomutase enzyme superfamily
journal, April 2013
- Lee, Yingying; Mehra-Chaudhary, Ritcha; Furdui, Cristina
- FEBS Journal, Vol. 280, Issue 11
Interpretation of ensembles created by multiple iterative rebuilding of macromolecular models.
text, January 2007
- Terwilliger, Thomas C.; Grosse-Kunstleve, Ralf W.; Afonine, Pavel V.
- Apollo - University of Cambridge Repository
Asp263 missense variants perturb the active site of human phosphoglucomutase 1
journal, February 2017
- Stiers, Kyle M.; Graham, Abigail C.; Kain, Bailee N.
- The FEBS Journal, Vol. 284, Issue 6
Real-space analysis of radiation-induced specific changes with independent component analysis
text, January 2018
- Borek, Dominika; Bromberg, Raquel; Hattne, Johan
- International Union of Crystallography (IUCr)
PHENIX: a comprehensive Python-based system for macromolecular structure solution.
text, January 2010
- Adams, Paul D.; Afonine, Pavel V.; Bunkóczi, Gábor
- Apollo - University of Cambridge Repository
Time-resolved x-ray diffraction reveals multiple conformations in the M-N transition of the bacteriorhodopsin photocycle
journal, December 2000
- Oka, T.; Yagi, N.; Fujisawa, T.
- Proceedings of the National Academy of Sciences, Vol. 97, Issue 26
Phosphorylation in the Catalytic Cleft Stabilizes and Attracts Domains of a Phosphohexomutase
journal, January 2015
- Xu, Jia; Lee, Yingying; Beamer, Lesa J.
- Biophysical Journal, Vol. 108, Issue 2
On vital aid: the why, what and how of validation
journal, January 2009
- Kleywegt, Gerard J.
- Acta Crystallographica Section D Biological Crystallography, Vol. 65, Issue 2
Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins
book, January 2013
- David, Charles C.; Jacobs, Donald J.
- Protein Dynamics
Kinetic Mechanism and pH Dependence of the Kinetic Parameters of Pseudomonas aeruginosa Phosphomannomutase/Phosphoglucomutase
journal, December 2001
- Naught, Laura E.; Tipton, Peter A.
- Archives of Biochemistry and Biophysics, Vol. 396, Issue 1
Evolutionary trace analysis of the α-D-phosphohexomutase superfamily
journal, August 2004
- Shackelford, Grant S.; Regni, Catherine A.; Beamer, Lesa J.
- Protein Science, Vol. 13, Issue 8
Protein flexibility: coordinate uncertainties and interpretation of structural differences
journal, October 2009
- Rashin, Alexander A.; Rashin, Abraham H. L.; Jernigan, Robert L.
- Acta Crystallographica Section D Biological Crystallography, Vol. 65, Issue 11
XDS
journal, January 2010
- Kabsch, Wolfgang
- Acta Crystallographica Section D Biological Crystallography, Vol. 66, Issue 2
Promotion of Enzyme Flexibility by Dephosphorylation and Coupling to the Catalytic Mechanism of a Phosphohexomutase
journal, January 2014
- Lee, Yingying; Villar, Maria T.; Artigues, Antonio
- Journal of Biological Chemistry, Vol. 289, Issue 8
Conservation of Functionally Important Global Motions in an Enzyme Superfamily across Varying Quaternary Structures
journal, November 2012
- Luebbering, Emily K.; Mick, Jacob; Singh, Ranjan K.
- Journal of Molecular Biology, Vol. 423, Issue 5
Exposing Hidden Alternative Backbone Conformations in X-ray Crystallography Using qFit
journal, October 2015
- Keedy, Daniel A.; Fraser, James S.; van den Bedem, Henry
- PLOS Computational Biology, Vol. 11, Issue 10
Formation and Reorientation of Glucose 1,6-Bisphosphate in the PMM/PGM Reaction: Transient-State Kinetic Studies †
journal, May 2005
- Naught, Laura E.; Tipton, Peter A.
- Biochemistry, Vol. 44, Issue 18
Evolution of oligomeric state through geometric coupling of protein interfaces
journal, May 2012
- Perica, T.; Chothia, C.; Teichmann, S. A.
- Proceedings of the National Academy of Sciences, Vol. 109, Issue 21
Some aspects of quantitative analysis and correction of radiation damage
journal, December 2005
- Diederichs, Kay
- Acta Crystallographica Section D Biological Crystallography, Vol. 62, Issue 1
High-Throughput Crystallography: Reliable and Efficient Identification of Fragment Hits
journal, August 2016
- Schiebel, Johannes; Krimmer, Stefan G.; Röwer, Karine
- Structure, Vol. 24, Issue 8
Application of Singular Value Decomposition to the Analysis of Time-Resolved Macromolecular X-Ray Data
journal, March 2003
- Schmidt, Marius; Rajagopal, Sudarshan; Ren, Zhong
- Biophysical Journal, Vol. 84, Issue 3
Phosphorylation-Dependent Effects on the Structural Flexibility of Phosphoglucosamine Mutase from Bacillus anthracis
journal, November 2017
- Stiers, Kyle M.; Xu, Jia; Lee, Yingying
- ACS Omega, Vol. 2, Issue 11
Quaternary structure, conformational variability and global motions of phosphoglucosamine mutase: Structure and motions of phosphoglucosamine mutase
journal, August 2011
- Mehra-Chaudhary, Ritcha; Mick, Jacob; Tanner, John J.
- FEBS Journal, Vol. 278, Issue 18
The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase
journal, October 2016
- Godsey, Michael H.; Davulcu, Omar; Nix, Jay C.
- Structure, Vol. 24, Issue 10
Sequence-structure relationships, expression profiles, and disease-associated mutations in the paralogs of phosphoglucomutase 1
journal, August 2017
- Muenks, Andrew G.; Stiers, Kyle M.; Beamer, Lesa J.
- PLOS ONE, Vol. 12, Issue 8
MolProbity : all-atom structure validation for macromolecular crystallography
journal, December 2009
- Chen, Vincent B.; Arendall, W. Bryan; Headd, Jeffrey J.
- Acta Crystallographica Section D Biological Crystallography, Vol. 66, Issue 1
Coot model-building tools for molecular graphics
journal, November 2004
- Emsley, Paul; Cowtan, Kevin
- Acta Crystallographica Section D Biological Crystallography, Vol. 60, Issue 12, p. 2126-2132
A method for the analysis of domain movements in large biomolecular complexes
journal, July 2009
- Poornam, Guru Prasad; Matsumoto, Atsushi; Ishida, Hisashi
- Proteins: Structure, Function, and Bioinformatics, Vol. 76, Issue 1
Trendspotting in the Protein Data Bank
journal, January 2013
- Berman, Helen M.; Coimbatore Narayanan, Buvaneswari; Costanzo, Luigi Di
- FEBS Letters, Vol. 587, Issue 8
Resolution of structural heterogeneity in dynamic crystallography
journal, May 2013
- Ren, Zhong; Chan, Peter W. Y.; Moffat, Keith
- Acta Crystallographica Section D Biological Crystallography, Vol. 69, Issue 6
Principal component analysis of native ensembles of biomolecular structures (PCA_NEST): insights into functional dynamics
journal, January 2009
- Yang, Lee-Wei; Eyal, Eran; Bahar, Ivet
- Bioinformatics, Vol. 25, Issue 5
Bio3d: an R package for the comparative analysis of protein structures
journal, August 2006
- Grant, B. J.; Rodrigues, A. P. C.; ElSawy, K. M.
- Bioinformatics, Vol. 22, Issue 21
Structural Basis of Diverse Substrate Recognition by the Enzyme PMM/PGM from P. aeruginosa
journal, March 2004
- Regni, Catherine; Naught, Laura; Tipton, Peter A.
- Structure, Vol. 12, Issue 1
PHENIX: a comprehensive Python-based system for macromolecular structure solution
journal, January 2010
- Adams, Paul D.; Afonine, Pavel V.; Bunkóczi, Gábor
- Acta Crystallographica Section D Biological Crystallography, Vol. 66, Issue 2, p. 213-221
Evolution of oligomeric state through geometric coupling of protein interfaces
journal, May 2012
- Perica, T.; Chothia, C.; Teichmann, S. A.
- Proceedings of the National Academy of Sciences, Vol. 109, Issue 21
Principal component analysis of native ensembles of biomolecular structures (PCA_NEST): insights into functional dynamics
journal, August 2009
- Yang, L. -W.; Eyal, E.; Bahar, I.
- Bioinformatics, Vol. 25, Issue 16
Mechanistic Insights on Human Phosphoglucomutase Revealed by Transition Path Sampling and Molecular Dynamics Calculations
journal, January 2018
- Brás, Natércia F.; Fernandes, Pedro A.; Ramos, Maria J.
- Chemistry - A European Journal, Vol. 24, Issue 8
A sampling problem in molecular dynamics simulations of macromolecules.
journal, April 1995
- Clarage, J. B.; Romo, T.; Andrews, B. K.
- Proceedings of the National Academy of Sciences, Vol. 92, Issue 8
Role of Conformational Dynamics in the Evolution of Retro-Aldolase Activity
journal, November 2017
- Romero-Rivera, Adrian; Garcia-Borràs, Marc; Osuna, Sílvia
- ACS Catalysis, Vol. 7, Issue 12
Phosphorylation in the Catalytic Cleft Stabilizes and Attracts Domains of a Phosphohexomutase
journal, January 2015
- Xu, Jia; Lee, Yingying; Beamer, Lesa J.
- Biophysical Journal, Vol. 108, Issue 2
Identification of an essential active-site residue in the α- d -phosphohexomutase enzyme superfamily
journal, April 2013
- Lee, Yingying; Mehra-Chaudhary, Ritcha; Furdui, Cristina
- FEBS Journal, Vol. 280, Issue 11
Linking Crystallographic Model and Data Quality
journal, May 2012
- Karplus, P. A.; Diederichs, K.
- Science, Vol. 336, Issue 6084
Interpretation of ensembles created by multiple iterative rebuilding of macromolecular models.
text, January 2007
- Terwilliger, Thomas C.; Grosse-Kunstleve, Ralf W.; Afonine, Pavel V.
- Apollo - University of Cambridge Repository
Domain-Opening and Dynamic Coupling in the α-Subunit of Heterotrimeric G Proteins
journal, July 2013
- Yao, Xin-Qiu; Grant, Barry J.
- Biophysical Journal, Vol. 105, Issue 2
PHENIX: a comprehensive Python-based system for macromolecular structure solution.
text, January 2010
- Adams, Paul D.; Afonine, Pavel V.; Bunkóczi, Gábor
- Apollo - University of Cambridge Repository
Formation and Reorientation of Glucose 1,6-Bisphosphate in the PMM/PGM Reaction: Transient-State Kinetic Studies †
journal, May 2005
- Naught, Laura E.; Tipton, Peter A.
- Biochemistry, Vol. 44, Issue 18
Microfocus diffraction from different regions of a protein crystal: structural variations and unit-cell polymorphism
journal, April 2018
- Thompson, Michael C.; Cascio, Duilio; Yeates, Todd O.
- Acta Crystallographica Section D Structural Biology, Vol. 74, Issue 5
Asp263 missense variants perturb the active site of human phosphoglucomutase 1
journal, February 2017
- Stiers, Kyle M.; Graham, Abigail C.; Kain, Bailee N.
- The FEBS Journal, Vol. 284, Issue 6
Role of Conformational Dynamics in the Evolution of Retro-Aldolase Activity
journal, November 2017
- Romero-Rivera, Adrian; Garcia-Borràs, Marc; Osuna, Sílvia
- ACS Catalysis, Vol. 7, Issue 12
Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen
journal, August 2020
- Zacarias, Sónia; Temporão, Adriana; Carpentier, Philippe
- JBIC Journal of Biological Inorganic Chemistry, Vol. 25, Issue 6
Evolutionary trace analysis of the α-D-phosphohexomutase superfamily
journal, August 2004
- Shackelford, Grant S.; Regni, Catherine A.; Beamer, Lesa J.
- Protein Science, Vol. 13, Issue 8
How good are my data and what is the resolution?
journal, June 2013
- Evans, Philip R.; Murshudov, Garib N.
- Acta Crystallographica Section D Biological Crystallography, Vol. 69, Issue 7
Accessing protein conformational ensembles using room-temperature X-ray crystallography
journal, September 2011
- Fraser, J. S.; van den Bedem, H.; Samelson, A. J.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 39
Kinetic Mechanism and pH Dependence of the Kinetic Parameters of Pseudomonas aeruginosa Phosphomannomutase/Phosphoglucomutase
journal, December 2001
- Naught, Laura E.; Tipton, Peter A.
- Archives of Biochemistry and Biophysics, Vol. 396, Issue 1
Trendspotting in the Protein Data Bank
journal, January 2013
- Berman, Helen M.; Coimbatore Narayanan, Buvaneswari; Costanzo, Luigi Di
- FEBS Letters, Vol. 587, Issue 8
Principal component analysis of native ensembles of biomolecular structures (PCA_NEST): insights into functional dynamics
journal, August 2009
- Yang, L. -W.; Eyal, E.; Bahar, I.
- Bioinformatics, Vol. 25, Issue 16
Residue mutations and their impact on protein structure and function: detecting beneficial and pathogenic changes
journal, January 2013
- Studer, Romain A.; Dessailly, Benoit H.; Orengo, Christine A.
- Biochemical Journal, Vol. 449, Issue 3
Time-resolved x-ray diffraction reveals multiple conformations in the M-N transition of the bacteriorhodopsin photocycle
journal, December 2000
- Oka, T.; Yagi, N.; Fujisawa, T.
- Proceedings of the National Academy of Sciences, Vol. 97, Issue 26
Remarks about protein structure precision
journal, March 1999
- Cruickshank, D. W. J.
- Acta Crystallographica Section D Biological Crystallography, Vol. 55, Issue 3
The Landscape of the Prion Protein's Structural Response to Mutation Revealed by Principal Component Analysis of Multiple NMR Ensembles
journal, August 2012
- Gendoo, Deena M. A.; Harrison, Paul M.
- PLoS Computational Biology, Vol. 8, Issue 8
Global indicators of X-ray data quality
journal, April 2001
- Weiss, Manfred S.
- Journal of Applied Crystallography, Vol. 34, Issue 2
Real-space analysis of radiation-induced specific changes with independent component analysis
text, January 2018
- Borek, Dominika; Bromberg, Raquel; Hattne, Johan
- International Union of Crystallography (IUCr)
Structural and functional characterization of the phosphoglucomutase from Xanthomonas citri subsp. citri
journal, December 2016
- Goto, Leandro Seiji; Vessoni Alexandrino, André; Malvessi Pereira, Camila
- Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, Vol. 1864, Issue 12
High-Throughput Crystallography: Reliable and Efficient Identification of Fragment Hits
journal, August 2016
- Schiebel, Johannes; Krimmer, Stefan G.; Röwer, Karine
- Structure, Vol. 24, Issue 8
Structural and functional characterization of the phosphoglucomutase from Xanthomonas citri subsp. citri
journal, December 2016
- Goto, Leandro Seiji; Vessoni Alexandrino, André; Malvessi Pereira, Camila
- Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, Vol. 1864, Issue 12
Interpretation of ensembles created by multiple iterative rebuilding of macromolecular models
journal, April 2007
- Terwilliger, Thomas C.; Grosse-Kunstleve, Ralf W.; Afonine, Pavel V.
- Acta Crystallographica Section D Biological Crystallography, Vol. 63, Issue 5
Polder maps: improving OMIT maps by excluding bulk solvent
journal, February 2017
- Liebschner, Dorothee; Afonine, Pavel V.; Moriarty, Nigel W.
- Acta Crystallographica Section D Structural Biology, Vol. 73, Issue 2, p. 148-157
A sampling problem in molecular dynamics simulations of macromolecules.
journal, April 1995
- Clarage, J. B.; Romo, T.; Andrews, B. K.
- Proceedings of the National Academy of Sciences, Vol. 92, Issue 8
Real-space analysis of radiation-induced specific changes with independent component analysis
journal, February 2018
- Borek, Dominika; Bromberg, Raquel; Hattne, Johan
- Journal of Synchrotron Radiation, Vol. 25, Issue 2
Cavin1 intrinsically disordered domains are essential for fuzzy electrostatic interactions and caveola formation
journal, February 2021
- Tillu, Vikas A.; Rae, James; Gao, Ya
- Nature Communications, Vol. 12, Issue 1
New software for the singular value decomposition of time-resolved crystallographic data
journal, June 2009
- Zhao, Yi; Schmidt, Marius
- Journal of Applied Crystallography, Vol. 42, Issue 4
Crystal Structure of PMM/PGM
journal, February 2002
- Regni, Catherine; Tipton, Peter A.; Beamer, Lesa J.
- Structure, Vol. 10, Issue 2
Phosphorylation-Dependent Effects on the Structural Flexibility of Phosphoglucosamine Mutase from Bacillus anthracis
journal, November 2017
- Stiers, Kyle M.; Xu, Jia; Lee, Yingying
- ACS Omega, Vol. 2, Issue 11
The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase
journal, October 2016
- Godsey, Michael H.; Davulcu, Omar; Nix, Jay C.
- Structure, Vol. 24, Issue 10
A graphical user interface to the CCP 4 program suite
journal, June 2003
- Potterton, Elizabeth; Briggs, Peter; Turkenburg, Maria
- Acta Crystallographica Section D Biological Crystallography, Vol. 59, Issue 7
Crystal Structures of N -Acetylglucosamine-phosphate Mutase, a Member of the α-d-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes
journal, May 2006
- Nishitani, Yuichi; Maruyama, Daisuke; Nonaka, Tsuyoshi
- Journal of Biological Chemistry, Vol. 281, Issue 28
Automated identification of functional dynamic contact networks from X-ray crystallography
journal, August 2013
- van den Bedem, Henry; Bhabha, Gira; Yang, Kun
- Nature Methods, Vol. 10, Issue 9
A method for the analysis of domain movements in large biomolecular complexes
journal, July 2009
- Poornam, Guru Prasad; Matsumoto, Atsushi; Ishida, Hisashi
- Proteins: Structure, Function, and Bioinformatics, Vol. 76, Issue 1
Sequence-structure relationships, expression profiles, and disease-associated mutations in the paralogs of phosphoglucomutase 1
journal, August 2017
- Muenks, Andrew G.; Stiers, Kyle M.; Beamer, Lesa J.
- PLOS ONE, Vol. 12, Issue 8
Compromised Catalysis and Potential Folding Defects in in Vitro Studies of Missense Mutants Associated with Hereditary Phosphoglucomutase 1 Deficiency
journal, October 2014
- Lee, Yingying; Stiers, Kyle M.; Kain, Bailee N.
- Journal of Biological Chemistry, Vol. 289, Issue 46
Accessing protein conformational ensembles using room-temperature X-ray crystallography
journal, September 2011
- Fraser, J. S.; van den Bedem, H.; Samelson, A. J.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 39
Quaternary structure, conformational variability and global motions of phosphoglucosamine mutase: Structure and motions of phosphoglucosamine mutase
journal, August 2011
- Mehra-Chaudhary, Ritcha; Mick, Jacob; Tanner, John J.
- FEBS Journal, Vol. 278, Issue 18
Domain-Opening and Dynamic Coupling in the α-Subunit of Heterotrimeric G Proteins
journal, July 2013
- Yao, Xin-Qiu; Grant, Barry J.
- Biophysical Journal, Vol. 105, Issue 2
Structural Basis of Diverse Substrate Recognition by the Enzyme PMM/PGM from P. aeruginosa
journal, March 2004
- Regni, Catherine; Naught, Laura; Tipton, Peter A.
- Structure, Vol. 12, Issue 1
Programmable RNA targeting by bacterial Argonaute nucleases with unconventional guide binding and cleavage specificity
journal, August 2022
- Lisitskaya, Lidiya; Shin, Yeonoh; Agapov, Aleksei
- Nature Communications, Vol. 13, Issue 1
The Reaction of Phosphohexomutase from Pseudomonas aeruginosa : STRUCTURAL INSIGHTS INTO A SIMPLE PROCESSIVE ENZYME
journal, April 2006
- Regni, Catherine; Schramm, Andrew M.; Beamer, Lesa J.
- Journal of Biological Chemistry, Vol. 281, Issue 22
A Kinetic Study of the Phosphoglucomutase Pathway
journal, April 1964
- Ray, William J.; Roscelli, Gertrude A.
- Journal of Biological Chemistry, Vol. 239, Issue 4
Works referencing / citing this record:
Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the α-d-phosphohexomutase superfamily
journal, March 2020
- Backe, Paul Hoff; Laerdahl, Jon K.; Kittelsen, Lene Svendsen
- Scientific Reports, Vol. 10, Issue 1
Mix-and-inject XFEL crystallography reveals gated conformational dynamics during enzyme catalysis
journal, December 2019
- Dasgupta, Medhanjali; Budday, Dominik; de Oliveira, Saulo H. P.
- Proceedings of the National Academy of Sciences, Vol. 116, Issue 51
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal