Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models
Abstract
We are developing a laboratory-scale model to improve our understanding and capacity to assess the biological risks of genetically engineered bacteria and their genetic elements in the natural environment. Our hypothetical scenario concerns an industrial bioreactor failure resulting in the introduction of genetically engineered bacteria to a downstream municipal wastewater treatment plant (MWWTP). As the first step towards developing a model for this scenario, we sampled microbial communities from the aeration basin of a MWWTP at three seasonal time points. Having established a baseline for community composition, we investigated how the community changed when propagated in the laboratory, including cell culture media conditions that could provide selective pressure in future studies. Specifically, using PhyloChip 16S-rRNA-gene targeting microarrays, we compared the compositions of sampled communities to those of inocula propagated in the laboratory in simulated wastewater conditionally amended with various carbon sources (glucose, chloroacetate, D-threonine) or the ionic liquid 1-ethyl-3-methylimidazolium chloride ([C2mim]Cl). Proteobacteria, Bacteroidetes, and Actinobacteria were predominant in both aeration basin and laboratory-cultured communities. Laboratory-cultured communities were enriched in γ-Proteobacteria. Enterobacteriaceae, and Aeromonadaceae were enriched by glucose, Pseudomonadaceae by chloroacetate and D-threonine, and Burkholderiacea by high (50 mM) concentrations of chloroacetate. Microbial communities cultured with chloroacetate and D-threonine were moremore »
- Authors:
-
- Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States)
- Publication Date:
- Research Org.:
- Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States)
- Sponsoring Org.:
- USDOE National Nuclear Security Administration (NNSA), Office of Defense Nuclear Nonproliferation
- OSTI Identifier:
- 1511406
- Grant/Contract Number:
- AC02-05CH11231
- Resource Type:
- Accepted Manuscript
- Journal Name:
- PLoS ONE
- Additional Journal Information:
- Journal Volume: 9; Journal Issue: 8; Journal ID: ISSN 1932-6203
- Publisher:
- Public Library of Science
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES
Citation Formats
Lee, Sangwon, Geller, Jil T., Torok, Tamas, Wu, Cindy H., Singer, Mary, Reid, Francine C., Tarjan, Daniel R., Hazen, Terry C., Arkin, Adam P., and Hillson, Nathan J. Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models. United States: N. p., 2014.
Web. doi:10.1371/journal.pone.0105689.
Lee, Sangwon, Geller, Jil T., Torok, Tamas, Wu, Cindy H., Singer, Mary, Reid, Francine C., Tarjan, Daniel R., Hazen, Terry C., Arkin, Adam P., & Hillson, Nathan J. Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models. United States. https://doi.org/10.1371/journal.pone.0105689
Lee, Sangwon, Geller, Jil T., Torok, Tamas, Wu, Cindy H., Singer, Mary, Reid, Francine C., Tarjan, Daniel R., Hazen, Terry C., Arkin, Adam P., and Hillson, Nathan J. Fri .
"Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models". United States. https://doi.org/10.1371/journal.pone.0105689. https://www.osti.gov/servlets/purl/1511406.
@article{osti_1511406,
title = {Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models},
author = {Lee, Sangwon and Geller, Jil T. and Torok, Tamas and Wu, Cindy H. and Singer, Mary and Reid, Francine C. and Tarjan, Daniel R. and Hazen, Terry C. and Arkin, Adam P. and Hillson, Nathan J.},
abstractNote = {We are developing a laboratory-scale model to improve our understanding and capacity to assess the biological risks of genetically engineered bacteria and their genetic elements in the natural environment. Our hypothetical scenario concerns an industrial bioreactor failure resulting in the introduction of genetically engineered bacteria to a downstream municipal wastewater treatment plant (MWWTP). As the first step towards developing a model for this scenario, we sampled microbial communities from the aeration basin of a MWWTP at three seasonal time points. Having established a baseline for community composition, we investigated how the community changed when propagated in the laboratory, including cell culture media conditions that could provide selective pressure in future studies. Specifically, using PhyloChip 16S-rRNA-gene targeting microarrays, we compared the compositions of sampled communities to those of inocula propagated in the laboratory in simulated wastewater conditionally amended with various carbon sources (glucose, chloroacetate, D-threonine) or the ionic liquid 1-ethyl-3-methylimidazolium chloride ([C2mim]Cl). Proteobacteria, Bacteroidetes, and Actinobacteria were predominant in both aeration basin and laboratory-cultured communities. Laboratory-cultured communities were enriched in γ-Proteobacteria. Enterobacteriaceae, and Aeromonadaceae were enriched by glucose, Pseudomonadaceae by chloroacetate and D-threonine, and Burkholderiacea by high (50 mM) concentrations of chloroacetate. Microbial communities cultured with chloroacetate and D-threonine were more similar to sampled field communities than those cultured with glucose or [C2mim]Cl. Although observed relative richness in operational taxonomic units (OTUs) was lower for laboratory cultures than for field communities, both flask and reactor systems supported phylogenetically diverse communities. These results importantly provide a foundation for laboratory models of industrial bioreactor failure scenarios.},
doi = {10.1371/journal.pone.0105689},
journal = {PLoS ONE},
number = 8,
volume = 9,
place = {United States},
year = {Fri Aug 22 00:00:00 EDT 2014},
month = {Fri Aug 22 00:00:00 EDT 2014}
}
Web of Science
Figures / Tables:
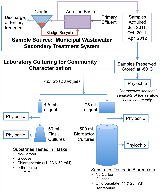 Figure 1: Overview of Experiments. All substrates were tested in triplicate, except for ‘No-Carbon in bioreactors’, which was tested in duplicate. The No-Carbon and [C2mim]Cl triplicates in flasks were pooled in order to provide adequate biomass for PhyloChip analysis.
Figure 1: Overview of Experiments. All substrates were tested in triplicate, except for ‘No-Carbon in bioreactors’, which was tested in duplicate. The No-Carbon and [C2mim]Cl triplicates in flasks were pooled in order to provide adequate biomass for PhyloChip analysis.
Works referenced in this record:
Crystal Structure of L-2-Haloacid Dehalogenase from Pseudomonas sp. YL : AN α/β HYDROLASE STRUCTURE THAT IS DIFFERENT FROM THE α/β HYDROLASE FOLD
journal, August 1996
- Hisano, Tamao; Hata, Yasuo; Fujii, Tomomi
- Journal of Biological Chemistry, Vol. 271, Issue 34
An auto-inducible mechanism for ionic liquid resistance in microbial biofuel production
journal, March 2014
- Ruegg, Thomas L.; Kim, Eun-Mi; Simmons, Blake A.
- Nature Communications, Vol. 5, Issue 1
GENETICALLY ENGINEERED ORGANISMS AND THE ENVIRONMENT: CURRENT STATUS AND RECOMMENDATIONS 1
journal, April 2005
- Snow, A. A.; Andow, D. A.; Gepts, P.
- Ecological Applications, Vol. 15, Issue 2
The use of rice seeds to produce human pharmaceuticals for oral therapy
journal, September 2013
- Wakasa, Yuhya; Takaiwa, Fumio
- Biotechnology Journal, Vol. 8, Issue 10
Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB
journal, July 2006
- DeSantis, T. Z.; Hugenholtz, P.; Larsen, N.
- Applied and Environmental Microbiology, Vol. 72, Issue 7, p. 5069-5072
Decontamination of a polychlorinated biphenyls-contaminated soil by phytoremediation-assisted bioaugmentation
journal, February 2013
- Secher, C.; Lollier, M.; Jézéquel, K.
- Biodegradation, Vol. 24, Issue 4
Synthetic biology: Tools to design microbes for the production of chemicals and fuels
journal, November 2013
- Seo, Sang Woo; Yang, Jina; Min, Byung Eun
- Biotechnology Advances, Vol. 31, Issue 6
Use of an Improved Statistical Method for Group Comparisons to Study Effects of Prairie Fire
journal, April 1985
- Zimmerman, Gregory M.; Goetz, Harold; Mielke, Paul W.
- Ecology, Vol. 66, Issue 2
The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases
journal, November 2011
- Caspi, R.; Altman, T.; Dreher, K.
- Nucleic Acids Research, Vol. 40, Issue D1
Genetics and Biochemistry of Dehalogenating Enzymes
journal, October 1994
- Janssen, D. B.; Pries, F.; Van der Ploeg, J. R.
- Annual Review of Microbiology, Vol. 48, Issue 1
Tackling the minority: sulfate-reducing bacteria in an archaea-dominated subsurface biofilm
journal, November 2012
- Probst, Alexander J.; Holman, Hoi-Ying N.; DeSantis, Todd Z.
- The ISME Journal, Vol. 7, Issue 3
High-density PhyloChip profiling of stimulated aquifer microbial communities reveals a complex response to acetate amendment
journal, April 2012
- Handley, Kim M.; Wrighton, Kelly C.; Piceno, Yvette M.
- FEMS Microbiology Ecology, Vol. 81, Issue 1
Cloning and Sequence Analysis of a Plasmid-encoded 2-Haloacid Dehalogenase Gene from Pseudomonas putida No. 109
journal, January 1994
- Kawasaki, Haruhiko; Toyama, Tohru; Maeda, Takuya
- Bioscience, Biotechnology, and Biochemistry, Vol. 58, Issue 1
Gene cloning and overproduction of low-specificity d -threonine aldolase from Alcaligenes xylosoxidans and its application for production of a key intermediate for parkinsonism drug
journal, July 2000
- Liu, J. Q.; Odani, M.; Yasuoka, T.
- Applied Microbiology and Biotechnology, Vol. 54, Issue 1
Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria
journal, May 2011
- Mendes, R.; Kruijt, M.; de Bruijn, I.
- Science, Vol. 332, Issue 6033
Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data
journal, August 2009
- Hamady, Micah; Lozupone, Catherine; Knight, Rob
- The ISME Journal, Vol. 4, Issue 1
Bacterial 2-haloacid dehalogenases: structures and reaction mechanisms
journal, October 2000
- Kurihara, Tatsuo; Esaki, Nobuyoshi; Soda, Kenji
- Journal of Molecular Catalysis B: Enzymatic, Vol. 10, Issue 1-3
Partial bioaugmentation to remove 3-chloroaniline slows bacterial species turnover rate in bioreactors
journal, December 2013
- Falk, Michael W.; Seshan, Hari; Dosoretz, Carlos
- Water Research, Vol. 47, Issue 19
Microbial Community Analysis of a Coastal Salt Marsh Affected by the Deepwater Horizon Oil Spill
journal, July 2012
- Beazley, Melanie J.; Martinez, Robert J.; Rajan, Suja
- PLoS ONE, Vol. 7, Issue 7
Microbial community dynamics in replicate membrane bioreactors – Natural reproducible fluctuations
journal, February 2009
- Falk, Michael W.; Song, Kyung-Guen; Matiasek, Michael G.
- Water Research, Vol. 43, Issue 3
Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation
journal, October 2006
- Letunic, I.; Bork, P.
- Bioinformatics, Vol. 23, Issue 1
FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments
journal, March 2010
- Price, Morgan N.; Dehal, Paramvir S.; Arkin, Adam P.
- PLoS ONE, Vol. 5, Issue 3
NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes
journal, July 2006
- DeSantis, T. Z.; Hugenholtz, P.; Keller, K.
- Nucleic Acids Research, Vol. 34, Issue Web Server
Environmentally relevant parameters affecting PCB degradation: carbon source- and growth phase-mitigated effects of the expression of the biphenyl pathway and associated genes in Burkholderia xenovorans LB400
journal, August 2009
- Parnell, J. Jacob; Denef, Vincent J.; Park, Joonhong
- Biodegradation, Vol. 21, Issue 1
Enzymatic Synthesis of β-Hydroxy-α-amino Acids Based on Recombinant d - and l -Threonine Aldolases
journal, December 1997
- Kimura, Teiji; Vassilev, Vassil P.; Shen, Gwo-Jenn
- Journal of the American Chemical Society, Vol. 119, Issue 49
Gene Expression Omnibus: NCBI gene expression and hybridization array data repository
journal, January 2002
- Edgar, R.
- Nucleic Acids Research, Vol. 30, Issue 1
Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli
journal, November 2011
- Bokinsky, G.; Peralta-Yahya, P. P.; George, A.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 50, p. 19949-19954
Metagenomic and Metatranscriptomic Analysis of Microbial Community Structure and Gene Expression of Activated Sludge
journal, May 2012
- Yu, Ke; Zhang, Tong
- PLoS ONE, Vol. 7, Issue 5
Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analysed by the 454-pyrosequencing technology
journal, August 2008
- Szczepanowski, Rafael; Bekel, Thomas; Goesmann, Alexander
- Journal of Biotechnology, Vol. 136, Issue 1-2
Rapid quantification and taxonomic classification of environmental DNA from both prokaryotic and eukaryotic origins using a microarray
journal, April 2005
- DeSantis, Todd Z.; Stone, Carol E.; Murray, Sonya R.
- FEMS Microbiology Letters, Vol. 245, Issue 2, p. 271-278
Four steps to avoid a synthetic-biology disaster
journal, February 2012
- Dana, Genya V.; Kuiken, Todd; Rejeski, David
- Nature, Vol. 483, Issue 7387
Deep-Sea Oil Plume Enriches Indigenous Oil-Degrading Bacteria
journal, August 2010
- Hazen, Terry C.; Dubinsky, Eric A.; DeSantis, Todd Z.
- Science, Vol. 330, Issue 6001
Substrate Specificity of Fluoroacetate Dehalogenase: An Insight from Crystallographic Analysis, Fluorescence Spectroscopy, and Theoretical Computations
journal, June 2012
- Nakayama, Tomonori; Kamachi, Takashi; Jitsumori, Keiji
- Chemistry - A European Journal, Vol. 18, Issue 27
Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications.
journal, January 1994
- Fetzner, S.; Lingens, F.
- Microbiological Reviews, Vol. 58, Issue 4
Substrate Specificity of Fluoroacetate Dehalogenase: An Insight from Crystallographic Analysis, Fluorescence Spectroscopy, and Theoretical Computations
journal, June 2012
- Nakayama, Tomonori; Kamachi, Takashi; Jitsumori, Keiji
- Chemistry - A European Journal, Vol. 18, Issue 27
Gene cloning and overproduction of low-specificity d -threonine aldolase from Alcaligenes xylosoxidans and its application for production of a key intermediate for parkinsonism drug
journal, July 2000
- Liu, J. Q.; Odani, M.; Yasuoka, T.
- Applied Microbiology and Biotechnology, Vol. 54, Issue 1
Environmentally relevant parameters affecting PCB degradation: carbon source- and growth phase-mitigated effects of the expression of the biphenyl pathway and associated genes in Burkholderia xenovorans LB400
journal, August 2009
- Parnell, J. Jacob; Denef, Vincent J.; Park, Joonhong
- Biodegradation, Vol. 21, Issue 1
Decontamination of a polychlorinated biphenyls-contaminated soil by phytoremediation-assisted bioaugmentation
journal, February 2013
- Secher, C.; Lollier, M.; Jézéquel, K.
- Biodegradation, Vol. 24, Issue 4
Synthetic biology: Tools to design microbes for the production of chemicals and fuels
journal, November 2013
- Seo, Sang Woo; Yang, Jina; Min, Byung Eun
- Biotechnology Advances, Vol. 31, Issue 6
Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analysed by the 454-pyrosequencing technology
journal, August 2008
- Szczepanowski, Rafael; Bekel, Thomas; Goesmann, Alexander
- Journal of Biotechnology, Vol. 136, Issue 1-2
Microbial community dynamics in replicate membrane bioreactors – Natural reproducible fluctuations
journal, February 2009
- Falk, Michael W.; Song, Kyung-Guen; Matiasek, Michael G.
- Water Research, Vol. 43, Issue 3
Partial bioaugmentation to remove 3-chloroaniline slows bacterial species turnover rate in bioreactors
journal, December 2013
- Falk, Michael W.; Seshan, Hari; Dosoretz, Carlos
- Water Research, Vol. 47, Issue 19
Substrate Specificity and Product Stereochemistry in the Dehalogenation of 2–Haloacids with the Crude Enzyme Preparation from Pseudomonas Putida
journal, April 1998
- Vyazmensky, Maria; Geresh, Shimona
- Enzyme and Microbial Technology, Vol. 22, Issue 5
Tackling the minority: sulfate-reducing bacteria in an archaea-dominated subsurface biofilm
journal, November 2012
- Probst, Alexander J.; Holman, Hoi-Ying N.; DeSantis, Todd Z.
- The ISME Journal, Vol. 7, Issue 3
An auto-inducible mechanism for ionic liquid resistance in microbial biofuel production
journal, March 2014
- Ruegg, Thomas L.; Kim, Eun-Mi; Simmons, Blake A.
- Nature Communications, Vol. 5, Issue 1
Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli
journal, November 2011
- Bokinsky, G.; Peralta-Yahya, P. P.; George, A.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 50, p. 19949-19954
R: A Language for Data Analysis and Graphics
journal, September 1996
- Ihaka, Ross; Gentleman, Robert
- Journal of Computational and Graphical Statistics, Vol. 5, Issue 3
Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation
journal, October 2006
- Letunic, I.; Bork, P.
- Bioinformatics, Vol. 23, Issue 1
Gene Expression Omnibus: NCBI gene expression and hybridization array data repository
journal, January 2002
- Edgar, R.
- Nucleic Acids Research, Vol. 30, Issue 1
NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes
journal, July 2006
- DeSantis, T. Z.; Hugenholtz, P.; Keller, K.
- Nucleic Acids Research, Vol. 34, Issue Web Server
Deep-Sea Oil Plume Enriches Indigenous Oil-Degrading Bacteria
journal, August 2010
- Hazen, Terry C.; Dubinsky, Eric A.; DeSantis, Todd Z.
- Science, Vol. 330, Issue 6001
Identification of Diverse Antimicrobial Resistance Determinants Carried on Bacterial, Plasmid, or Viral Metagenomes from an Activated Sludge Microbial Assemblage
journal, April 2010
- Parsley, L. C.; Consuegra, E. J.; Kakirde, K. S.
- Applied and Environmental Microbiology, Vol. 76, Issue 11
Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications
journal, December 1994
- Fetzner, S.; Lingens, F.
- Microbiological Reviews, Vol. 58, Issue 4
Genetics and Biochemistry of Dehalogenating Enzymes
journal, October 1994
- Janssen, D. B.; Pries, F.; Van der Ploeg, J. R.
- Annual Review of Microbiology, Vol. 48, Issue 1
FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments
journal, March 2010
- Price, Morgan N.; Dehal, Paramvir S.; Arkin, Adam P.
- PLoS ONE, Vol. 5, Issue 3
Metagenomic and Metatranscriptomic Analysis of Microbial Community Structure and Gene Expression of Activated Sludge
journal, May 2012
- Yu, Ke; Zhang, Tong
- PLoS ONE, Vol. 7, Issue 5
Microbial Community Analysis of a Coastal Salt Marsh Affected by the Deepwater Horizon Oil Spill
journal, July 2012
- Beazley, Melanie J.; Martinez, Robert J.; Rajan, Suja
- PLoS ONE, Vol. 7, Issue 7
Species Assemblages and Indicator Species: The Need for a Flexible Asymmetrical Approach
journal, August 1997
- Dufrene, Marc; Legendre, Pierre
- Ecological Monographs, Vol. 67, Issue 3
Preparing synthetic biology for the world
journal, January 2013
- Moe-Behrens, Gerd H. G.; Davis, Rene; Haynes, Karmella A.
- Frontiers in Microbiology, Vol. 4
Works referencing / citing this record:
Effect of hydraulic retention time on microbial community structure in wastewater treatment electro-bioreactors
journal, March 2018
- ElNaker, Nancy A.; Yousef, Ahmed F.; Hasan, Shadi W.
- MicrobiologyOpen, Vol. 7, Issue 4
Comparative diversity of microbiomes and Resistomes in beef feedlots, downstream environments and urban sewage influent
journal, August 2019
- Zaheer, Rahat; Lakin, Steven M.; Polo, Rodrigo Ortega
- BMC Microbiology, Vol. 19, Issue 1
Comparative diversity of microbiomes and Resistomes in beef feedlots, downstream environments and urban sewage influent
journal, August 2019
- Zaheer, Rahat; Lakin, Steven M.; Polo, Rodrigo Ortega
- BMC Microbiology, Vol. 19, Issue 1
Metagenomic analysis of an ecological wastewater treatment plant’s microbial communities and their potential to metabolize pharmaceuticals
journal, July 2016
- Balcom, Ian N.; Driscoll, Heather; Vincent, James
- F1000Research, Vol. 5
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal