Structural Evidence of a Major Conformational Change Triggered by Substrate Binding in DapE Enzymes: Impact on the Catalytic Mechanism
Abstract
In this paper, the X-ray crystal structure of the dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase from Haemophilus influenzae (HiDapE) bound by the products of hydrolysis, succinic acid and l,l-DAP, was determined at 1.95 Å. Surprisingly, the structure bound to the products revealed that HiDapE undergoes a significant conformational change in which the catalytic domain rotates ~50° and shifts ~10.1 Å (as measured at the position of the Zn atoms) relative to the dimerization domain. This heretofore unobserved closed conformation revealed significant movements within the catalytic domain compared to that of wild-type HiDapE, which results in effectively closing off access to the dinuclear Zn(II) active site with the succinate carboxylate moiety bridging the dinculear Zn(II) cluster in a μ-1,3 fashion forming a bis(μ-carboxylato)dizinc(II) core with a Zn–Zn distance of 3.8 Å. Surprisingly, His194.B, which is located on the dimerization domain of the opposing chain ~10.1 Å from the dinuclear Zn(II) active site, forms a hydrogen bond (2.9 Å) with the oxygen atom of succinic acid bound to Zn2, forming an oxyanion hole. As the closed structure forms upon substrate binding, the movement of His194.B by more than ~10 Å is critical, based on site-directed mutagenesis data, for activation of the scissile carbonyl carbonmore »
- Authors:
-
- Argonne National Lab. (ANL), Argonne, IL (United States). Midwest Center for Structural Genomics. Structural Biology Center. Biosciences Division
- Loyola Univ. Chicago, IL (United States). Dept. of Chemistry and Biochemistry
- Marquette Univ., Milwaukee, WI (United States). Dept. of Chemistry
- Publication Date:
- Research Org.:
- Argonne National Laboratory (ANL), Argonne, IL (United States); Loyola Univ. Chicago, IL (United States); Marquette Univ., Milwaukee, WI (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC), Biological and Environmental Research (BER); National Inst. of Health (NIH) (United States); National Science Foundation (NSF); Todd Wehr Foundation (United States)
- OSTI Identifier:
- 1427504
- Grant/Contract Number:
- AC02-06CH11357; HHSN272200700058C; HHSN272201200026C; CHE-1412443
- Resource Type:
- Accepted Manuscript
- Journal Name:
- Biochemistry
- Additional Journal Information:
- Journal Volume: 57; Journal Issue: 5; Journal ID: ISSN 0006-2960
- Publisher:
- American Chemical Society (ACS)
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES; 37 INORGANIC, ORGANIC, PHYSICAL, AND ANALYTICAL CHEMISTRY
Citation Formats
Nocek, Boguslaw, Reidl, Cory, Starus, Anna, Heath, Tahirah, Bienvenue, David, Osipiuk, Jerzy, Jedrzejczak, Robert, Joachimiak, Andrzej, Becker, Daniel P., and Holz, Richard C. Structural Evidence of a Major Conformational Change Triggered by Substrate Binding in DapE Enzymes: Impact on the Catalytic Mechanism. United States: N. p., 2017.
Web. doi:10.1021/acs.biochem.7b01151.
Nocek, Boguslaw, Reidl, Cory, Starus, Anna, Heath, Tahirah, Bienvenue, David, Osipiuk, Jerzy, Jedrzejczak, Robert, Joachimiak, Andrzej, Becker, Daniel P., & Holz, Richard C. Structural Evidence of a Major Conformational Change Triggered by Substrate Binding in DapE Enzymes: Impact on the Catalytic Mechanism. United States. https://doi.org/10.1021/acs.biochem.7b01151
Nocek, Boguslaw, Reidl, Cory, Starus, Anna, Heath, Tahirah, Bienvenue, David, Osipiuk, Jerzy, Jedrzejczak, Robert, Joachimiak, Andrzej, Becker, Daniel P., and Holz, Richard C. Fri .
"Structural Evidence of a Major Conformational Change Triggered by Substrate Binding in DapE Enzymes: Impact on the Catalytic Mechanism". United States. https://doi.org/10.1021/acs.biochem.7b01151. https://www.osti.gov/servlets/purl/1427504.
@article{osti_1427504,
title = {Structural Evidence of a Major Conformational Change Triggered by Substrate Binding in DapE Enzymes: Impact on the Catalytic Mechanism},
author = {Nocek, Boguslaw and Reidl, Cory and Starus, Anna and Heath, Tahirah and Bienvenue, David and Osipiuk, Jerzy and Jedrzejczak, Robert and Joachimiak, Andrzej and Becker, Daniel P. and Holz, Richard C.},
abstractNote = {In this paper, the X-ray crystal structure of the dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase from Haemophilus influenzae (HiDapE) bound by the products of hydrolysis, succinic acid and l,l-DAP, was determined at 1.95 Å. Surprisingly, the structure bound to the products revealed that HiDapE undergoes a significant conformational change in which the catalytic domain rotates ~50° and shifts ~10.1 Å (as measured at the position of the Zn atoms) relative to the dimerization domain. This heretofore unobserved closed conformation revealed significant movements within the catalytic domain compared to that of wild-type HiDapE, which results in effectively closing off access to the dinuclear Zn(II) active site with the succinate carboxylate moiety bridging the dinculear Zn(II) cluster in a μ-1,3 fashion forming a bis(μ-carboxylato)dizinc(II) core with a Zn–Zn distance of 3.8 Å. Surprisingly, His194.B, which is located on the dimerization domain of the opposing chain ~10.1 Å from the dinuclear Zn(II) active site, forms a hydrogen bond (2.9 Å) with the oxygen atom of succinic acid bound to Zn2, forming an oxyanion hole. As the closed structure forms upon substrate binding, the movement of His194.B by more than ~10 Å is critical, based on site-directed mutagenesis data, for activation of the scissile carbonyl carbon of the substrate for nucleophilic attack by a hydroxide nucleophile. Employing the HiDapE product-bound structure as the starting point, a reverse engineering approach called product-based transition-state modeling provided structural models for each major catalytic step. Finally, these data provide insight into the catalytic reaction mechanism and also the future design of new, potent inhibitors of DapE enzymes.},
doi = {10.1021/acs.biochem.7b01151},
journal = {Biochemistry},
number = 5,
volume = 57,
place = {United States},
year = {Fri Dec 22 00:00:00 EST 2017},
month = {Fri Dec 22 00:00:00 EST 2017}
}
Web of Science
Figures / Tables:
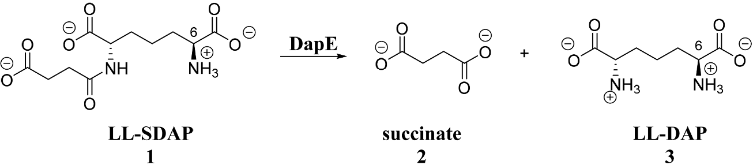 Figure 1: Enzymatic cleavage of L,L-SDAP (1) by DapE yielding succinate (2) and L,L-DAP (3).
Figure 1: Enzymatic cleavage of L,L-SDAP (1) by DapE yielding succinate (2) and L,L-DAP (3).
Works referenced in this record:
Antimicrobial resistance: action to combat the rising microbial challenges
journal, June 2013
- Paphitou, Niki I.
- International Journal of Antimicrobial Agents, Vol. 42
Antibiotics and Bacterial Resistance in the 21st Century
journal, January 2014
- Fair, Richard J.; Tor, Yitzhak
- Perspectives in Medicinal Chemistry, Vol. 6
Lysine biosynthesis in bacteria: a metallodesuccinylase as a potential antimicrobial target
journal, December 2012
- Gillner, Danuta M.; Becker, Daniel P.; Holz, Richard C.
- JBIC Journal of Biological Inorganic Chemistry, Vol. 18, Issue 2
Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis.
journal, January 1996
- Pavelka, M. S.; Jacobs, W. R.
- Journal of bacteriology, Vol. 178, Issue 22
Inhibition of lysine biosynthesis: an evolving antibiotic strategy
journal, January 2007
- Hutton, Craig A.; Perugini, Matthew A.; Gerrard, Juliet A.
- Molecular BioSystems, Vol. 3, Issue 7
The dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase from Haemophilus influenzae contains two active-site histidine residues
journal, August 2008
- Gillner, Danuta M.; Bienvenue, David L.; Nocek, Boguslaw P.
- JBIC Journal of Biological Inorganic Chemistry, Vol. 14, Issue 1
Structural Basis for Catalysis by the Mono- and Dimetalated Forms of the dapE-Encoded N-succinyl-l,l-Diaminopimelic Acid Desuccinylase
journal, April 2010
- Nocek, Boguslaw P.; Gillner, Danuta M.; Fan, Yao
- Journal of Molecular Biology, Vol. 397, Issue 3
The Dimerization Domain in DapE Enzymes Is required for Catalysis
journal, May 2014
- Nocek, Boguslaw; Starus, Anna; Makowska-Grzyska, Magdalena
- PLoS ONE, Vol. 9, Issue 5
Substrate Specificity, Metal Binding Properties, and Spectroscopic Characterization of the DapE-Encoded N-Succinyl-l ,l -Diaminopimelic Acid Desuccinylase from Haemophilus influenzae
journal, August 2003
- Bienvenue, David L.; Gilner, Danuta M.; Davis, Ryan S.
- Biochemistry, Vol. 42, Issue 36
Inhibition of the dapE -Encoded N -Succinyl- l , l -diaminopimelic Acid Desuccinylase from Neisseria meningitidis by l -Captopril
journal, July 2015
- Starus, Anna; Nocek, Boguslaw; Bennett, Brian
- Biochemistry, Vol. 54, Issue 31
[20] Processing of X-ray diffraction data collected in oscillation mode
book, January 1997
- Otwinowski, Zbyszek; Minor, Wladek
- Macromolecular Crystallography Part A, 307-326
Molecular replacement with MOLREP
journal, December 2009
- Vagin, Alexei; Teplyakov, Alexei
- Acta Crystallographica Section D Biological Crystallography, Vol. 66, Issue 1
REFMAC 5 for the refinement of macromolecular crystal structures
journal, March 2011
- Murshudov, Garib N.; Skubák, Pavol; Lebedev, Andrey A.
- Acta Crystallographica Section D Biological Crystallography, Vol. 67, Issue 4
PHENIX: a comprehensive Python-based system for macromolecular structure solution
journal, January 2010
- Adams, Paul D.; Afonine, Pavel V.; Bunkóczi, Gábor
- Acta Crystallographica Section D Biological Crystallography, Vol. 66, Issue 2, p. 213-221
MolProbity: all-atom contacts and structure validation for proteins and nucleic acids
journal, May 2007
- Davis, I. W.; Leaver-Fay, A.; Chen, V. B.
- Nucleic Acids Research, Vol. 35, Issue Web Server
The generalized Born/volume integral implicit solvent model: Estimation of the free energy of hydration using London dispersion instead of atomic surface area
journal, February 2008
- Labute, Paul
- Journal of Computational Chemistry, Vol. 29, Issue 10
Hydrolysis of N -Succinyl- l , l -diaminopimelic Acid by the Haemophilus influenzae dapE -Encoded Desuccinylase: Metal Activation, Solvent Isotope Effects, and Kinetic Mechanism †
journal, July 1998
- Born, Timothy L.; Zheng, Renjian; Blanchard, John S.
- Biochemistry, Vol. 37, Issue 29
Structural and mechanistic insight into substrate binding from the conformational dynamics in apo and substrate-bound DapE enzyme
journal, January 2016
- Dutta, Debodyuti; Mishra, Sabyashachi
- Physical Chemistry Chemical Physics, Vol. 18, Issue 3
Small revisions to predicted distances around metal sites in proteins
journal, May 2006
- Harding, Marjorie M.
- Acta Crystallographica Section D Biological Crystallography, Vol. 62, Issue 6
Structural Analysis of a Ternary Complex of Allantoate Amidohydrolase from Escherichia coli Reveals its Mechanics
journal, April 2007
- Agarwal, Rakhi; Burley, Stephen K.; Swaminathan, Subramanyam
- Journal of Molecular Biology, Vol. 368, Issue 2
Structural and Functional Analyses Reveal That Staphylococcus aureus Antibiotic Resistance Factor HmrA Is a Zinc-dependent Endopeptidase
journal, July 2011
- Botelho, Tiago O.; Guevara, Tibisay; Marrero, Aniebrys
- Journal of Biological Chemistry, Vol. 286, Issue 29
Crystallization and preliminary X-ray analysis of β-alanine synthase from the yeast Saccharomyces kluyveri
journal, June 2003
- Dobritzsch, Doreen; Gojković, Zoran; Andersen, Birgit
- Acta Crystallographica Section D Biological Crystallography, Vol. 59, Issue 7
Enzyme Catalysis by Hydrogen Bonds: The Balance between Transition State Binding and Substrate Binding in Oxyanion Holes
journal, March 2010
- Simón, Luis; Goodman, Jonathan M.
- The Journal of Organic Chemistry, Vol. 75, Issue 6
Solid-State 17 O NMR of Unstable Acyl-Enzyme Intermediates: A Direct Probe of Hydrogen Bonding Interactions in the Oxyanion Hole of Serine Proteases
journal, October 2016
- Tang, Aaron W.; Kong, Xianqi; Terskikh, Victor
- The Journal of Physical Chemistry B, Vol. 120, Issue 43
The d apE -encoded N -Succinyl- l , l -Diaminopimelic Acid Desuccinylase from Haemophilus i nfluenzae Is a Dinuclear Metallohydrolase
journal, December 2003
- Cosper, Nathaniel J.; Bienvenue, David L.; Shokes, Jacob E.
- Journal of the American Chemical Society, Vol. 125, Issue 48
Transition State Analog L-Leucinephosphonic Acid Bound to Bovine Lens Leucine Aminopeptidase: X-ray Structure at 1.65 .ANG. Resolution in a New Crystal Form
journal, July 1995
- Strater, Norbert; Lipscomb, William N.
- Biochemistry, Vol. 34, Issue 28
The structural and energetic aspects of substrate binding and the mechanism of action of the DapE-encoded N-succinyl- l , l -diaminopimelic acid desuccinylase (DapE) investigated using a hybrid QM/MM method
journal, January 2014
- Dutta, Debodyuti; Mishra, Sabyashachi
- Phys. Chem. Chem. Phys., Vol. 16, Issue 47
Kinetic and spectroscopic characterization of the E134A- and E134D-altered dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase from Haemophilus influenzae
journal, January 2006
- Davis, Ryan; Bienvenue, David; Swierczek, Sabina I.
- JBIC Journal of Biological Inorganic Chemistry, Vol. 11, Issue 2
Structural basis of catalysis by monometalated methionine aminopeptidase
journal, June 2006
- Ye, Q. -Z.; Xie, S. -X.; Ma, Z. -Q.
- Proceedings of the National Academy of Sciences, Vol. 103, Issue 25
Kinetic and Spectroscopic Characterization of the H178A Methionyl Aminopeptidase from Escherichia coli †
journal, May 2003
- Copik, Alicja J.; Swierczek, Sabina I.; Lowther, W. Todd
- Biochemistry, Vol. 42, Issue 20
The aminopeptidase from Aeromonas proteolytica: structure and mechanism of co-catalytic metal centers involved in peptide hydrolysis
journal, October 2002
- Holz, Richard C.
- Coordination Chemistry Reviews, Vol. 232, Issue 1-2
Works referencing / citing this record:
Practical spectrophotometric assay for the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase, a potential antibiotic target
journal, April 2018
- Heath, Tahirah K.; Lutz, Marlon R.; Reidl, Cory T.
- PLOS ONE, Vol. 13, Issue 4

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal