Collision-free poisson motion planning in ultra high-dimensional molecular conformation spaces
- Stanford Univ., CA (United States). Molecular and Cellular Physiology; SLAC National Accelerator Lab., Menlo Park, CA (United States). Bioscience Division
- Univ. of Erlangen-Nuremberg, Erlangen (Germany). Chair of Applied Dynamics
- SLAC National Accelerator Lab., Menlo Park, CA (United States). Bioscience Division
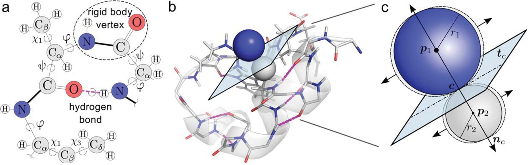
The function of protein, RNA, and DNA is modulated by fast, dynamic exchanges between three-dimensional conformations. Conformational sampling of biomolecules with exact and nullspace inverse kinematics, using rotatable bonds as revolute joints and noncovalent interactions as holonomic constraints, can accurately characterize these native ensembles. However, sampling biomolecules remains challenging owing to their ultra-high dimensional configuration spaces, and the requirement to avoid (self-) collisions, which results in low acceptance rates. In this paper, we present two novel mechanisms to overcome these limitations. First, we introduce temporary constraints between near-colliding links. The resulting constraint varieties instantaneously redirect the search for collision-free conformations, and couple motions between distant parts of the linkage. Second, we adapt a randomized Poisson-disk motion planner, which prevents local oversampling and widens the search, to ultra-high dimensions. Tests on several model systems show that the sampling acceptance rate can increase from 16% to 70%, and that the conformational coverage in loop modeling measured as average closeness to existing loop conformations doubled. Finally, correlated protein motions identified with our algorithm agree with those from MD simulations.
- Research Organization:
- SLAC National Accelerator Laboratory (SLAC), Menlo Park, CA (United States); Stanford Univ., CA (United States); Univ. of Erlangen-Nuremberg, Erlangen (Germany)
- Sponsoring Organization:
- USDOE; Deutsche Telekom Stiftung (Germany); Novo Nordisk Foundation (Denmark)
- Grant/Contract Number:
- AC02-76SF00515; NNF15OC0015268
- OSTI ID:
- 1471534
- Journal Information:
- Journal of Computational Chemistry, Vol. 39, Issue 12; ISSN 0192-8651
- Publisher:
- WileyCopyright Statement
- Country of Publication:
- United States
- Language:
- English
Web of Science
Similar Records
qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps
Similarity Downselection: Finding the n Most Dissimilar Molecular Conformers for Reference-Free Metabolomics