Improved assemblies using a source-agnostic pipeline for MetaGenomic Assembly by Merging (MeGAMerge) of contigs
- Los Alamos National Lab. (LANL), Los Alamos, NM (United States); USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States)
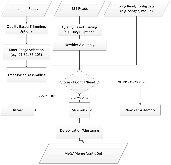
Assembly of metagenomic samples is a very complex process, with algorithms designed to address sequencing platform-specific issues, (read length, data volume, and/or community complexity), while also faced with genomes that differ greatly in nucleotide compositional biases and in abundance. To address these issues, we have developed a post-assembly process: MetaGenomic Assembly by Merging (MeGAMerge). We compare this process to the performance of several assemblers, using both real, and in-silico generated samples of different community composition and complexity. MeGAMerge consistently outperforms individual assembly methods, producing larger contigs with an increased number of predicted genes, without replication of data. MeGAMerge contigs are supported by read mapping and contig alignment data, when using synthetically-derived and real metagenomic data, as well as by gene prediction analyses and similarity searches. Ultimately, MeGAMerge is a flexible method that generates improved metagenome assemblies, with the ability to accommodate upcoming sequencing platforms, as well as present and future assembly algorithms.
- Research Organization:
- Los Alamos National Laboratory (LANL), Los Alamos, NM (United States)
- Sponsoring Organization:
- USDOE Office of Science (SC); U.S. Department of Homeland Security
- Grant/Contract Number:
- AC02-05CH11231; HSHQDC08X00790; B104153I; B084531I
- OSTI ID:
- 1259288
- Journal Information:
- Scientific Reports, Vol. 4; ISSN 2045-2322
- Publisher:
- Nature Publishing GroupCopyright Statement
- Country of Publication:
- United States
- Language:
- English
Web of Science
Similar Records
Use of simulated data sets to evaluate the fidelity of metagenomic processing methods
Benchmarking viromics: an in silico evaluation of metagenome-enabled estimates of viral community composition and diversity