
ID: growdb:01
Please cite this project and all data from these samples as:
Borton, et al. (2022) GROWdb US River Systems - Samples. [Data set]. DOE Systems Biology Knowledgebase. doi:10.25982/109073.30/1895615.
We developed the Genome Resolved Open Watersheds database (GROWdb), which aims to increase genomic sampling and understanding of global river microbiomes. An emphasis of GROWdb is to create a publicly available and ever-expanding microbial genome database that is focused on rivers while being interoperable with databases from other ecosystems. GROWdb is based on a network-of-networks approach to move beyond a small collection of well-studied rivers, towards a spatially distributed, global network of systematic observations. GROWdb represents the first microbial, river-focused resource parsed at various scales from genes to MAGs to community level including expression and potential based measurements that will be of interest to microbiologists, ecologists, geochemists, hydrologists, and modelers.
GROWdb contains data from various research campaigns, please acknowledge the following data generators, as appropriate:
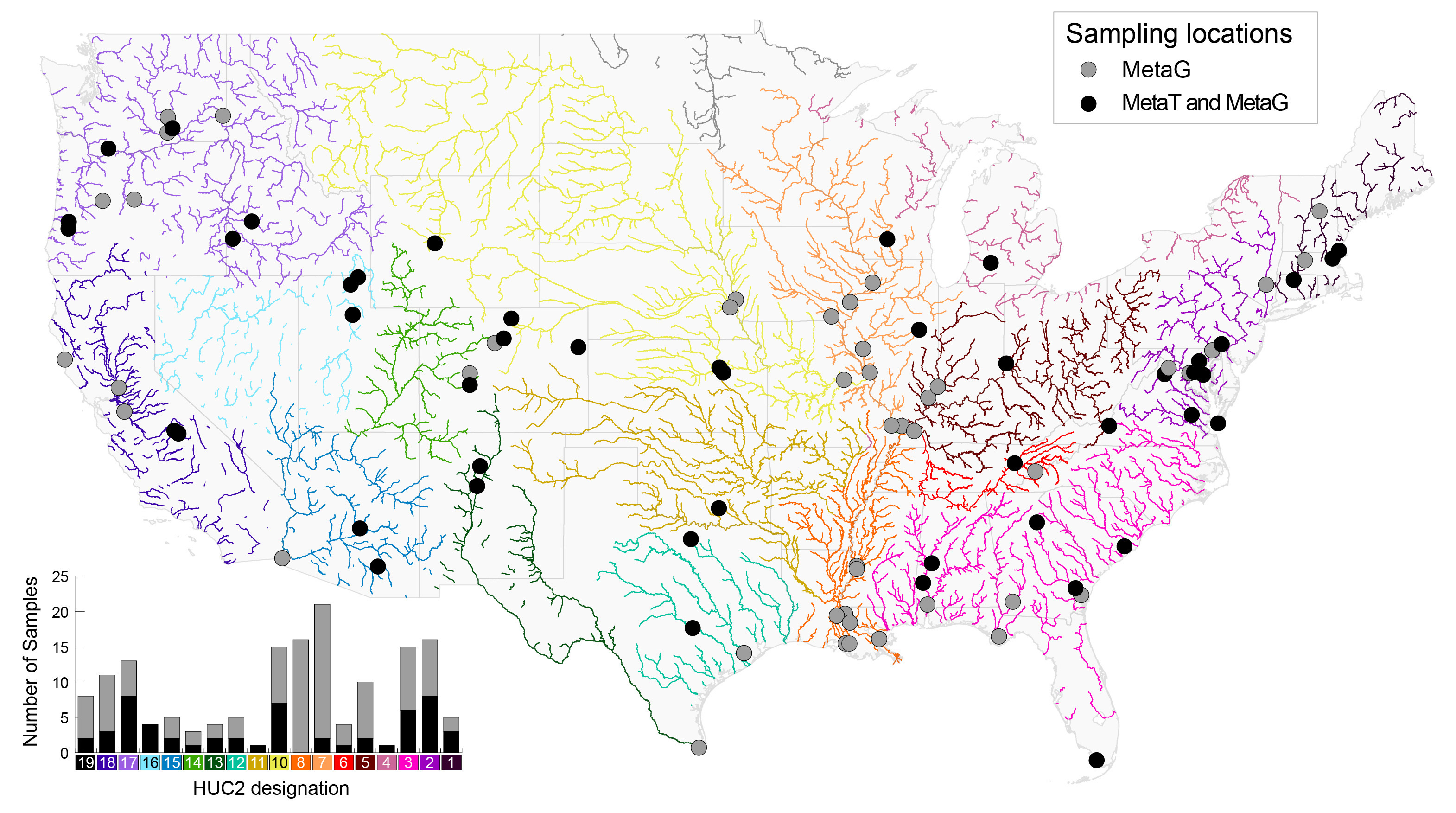
Total Samples loaded onto this Narrative: 178
Note: Not all GROW samples may be loaded into KBase
The data underlying GROWdb are accessible across various platforms to ensure all levels of data structure are widely available. First, all reads and MAGs are publicly hosted on National Center for Biotechnology (NCBI) under Bioproject PRJNA946291. Second, all data related data presented here including MAG annotations, extended data tables, phylogenetic tree files, antibiotic resistance gene database files, and MAG abundance tables are available in Zenodo (link).
Beyond the flat database files listed above, our aim for GROWdb was to maximize data use by making the data available in searchable and interactive platforms including the National Microbiome Data Collaborative (NMDC) data portal, the Department of Energy’s Systems Biology Knowledgebase (KBase), and a GROW specific user interface released here, GROWdb Explorer. Each platform provides different ways to interact with GROWdb:
Other linked narratives in KBase:
GROWdb Explorer GROWdb data is also explorable through a graphical user interface built through the Colorado State University Geospatial Centroid (https://geocentroid.shinyapps.io/GROWdatabase/), allowing users to search and graph microbial and spatial data simultaneously.
In summary, this microbial genome resource represents the first publicly available genome collection from rivers and offers data that can be leveraged across microbiome studies. GROWdb is an expanding repository to incorporate and unify global river multi-omic data for the future.
| Narrative Content | Info | Narrative Link | |
|---|---|---|---|
| GROW MAGs | Dereplicated MAGs 2093 | https://narrative.kbase.us/narrative/106867 | |
| GROW Metabolic Models | Models derived based on MAGs | https://narrative.kbase.us/narrative/107864 |