Unraveling Microbial Communities Associated with Methylmercury Production in Paddy Soils
Abstract
Here, we report rice consumption is now recognized as an important pathway of human exposure to the neurotoxin methylmercury (MeHg), particularly in countries where rice is a staple food. Although the discovery of a two-gene cluster hgcAB has linked Hg methylation to several phylogenetically diverse groups of anaerobic microorganisms converting inorganic mercury (Hg) to MeHg, the prevalence and diversity of Hg methylators in microbial communities of rice paddy soils remain unclear. We characterized the abundance and distribution of hgcAB genes using third-generation PacBio long-read sequencing and Illumina short-read metagenomic sequencing, in combination with quantitative PCR analyses in several mine-impacted paddy soils from southwest China. Both Illumina and PacBio sequencing analyses revealed that Hg methylating communities were dominated by iron-reducing bacteria (i.e., Geobacter) and methanogens, with a relatively low abundance of hgcA+ sulfate-reducing bacteria in the soil. A positive correlation was observed between the MeHg content in soil and the relative abundance of Geobacter carrying the hgcA gene. Phylogenetic analysis also uncovered some hgcAB sequences closely related to three novel Hg methylators, Geobacter anodireducens, Desulfuromonas sp. DDH964, and Desulfovibrio sp. J2, among which G. anodireducens was validated for its ability to methylate Hg. Lastly, these findings shed new light on microbialmore »
- Authors:
-
- Huazhong Agricultural University, Wuhan (China); Chinese Academy of Sciences (CAS), Beijing (China)
- Oak Ridge National Lab. (ORNL), Oak Ridge, TN (United States)
- Chinese Academy of Sciences (CAS), Beijing (China)
- The University of Melbourne, Victoria (Australia)
- Zhejiang Univ. (China)
- Chinese Academy of Sciences (CAS), Beijing (China); The University of Melbourne, Victoria (Australia)
- Oak Ridge National Lab. (ORNL), Oak Ridge, TN (United States); Univ. of Tennessee, Knoxville, TN (United States)
- Publication Date:
- Research Org.:
- Oak Ridge National Laboratory (ORNL), Oak Ridge, TN (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC), Biological and Environmental Research (BER)
- OSTI Identifier:
- 1490597
- Grant/Contract Number:
- AC05-00OR22725
- Resource Type:
- Accepted Manuscript
- Journal Name:
- Environmental Science and Technology
- Additional Journal Information:
- Journal Volume: 52; Journal Issue: 22; Journal ID: ISSN 0013-936X
- Publisher:
- American Chemical Society (ACS)
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES; 54 ENVIRONMENTAL SCIENCES
Citation Formats
Liu, Yu-Rong, Johs, Alexander, Bi, Li, Lu, Xia, Hu, Hang-Wei, Sun, Dan, He, Ji-Zheng, and Gu, Baohua. Unraveling Microbial Communities Associated with Methylmercury Production in Paddy Soils. United States: N. p., 2018.
Web. doi:10.1021/acs.est.8b03052.
Liu, Yu-Rong, Johs, Alexander, Bi, Li, Lu, Xia, Hu, Hang-Wei, Sun, Dan, He, Ji-Zheng, & Gu, Baohua. Unraveling Microbial Communities Associated with Methylmercury Production in Paddy Soils. United States. https://doi.org/10.1021/acs.est.8b03052
Liu, Yu-Rong, Johs, Alexander, Bi, Li, Lu, Xia, Hu, Hang-Wei, Sun, Dan, He, Ji-Zheng, and Gu, Baohua. Wed .
"Unraveling Microbial Communities Associated with Methylmercury Production in Paddy Soils". United States. https://doi.org/10.1021/acs.est.8b03052. https://www.osti.gov/servlets/purl/1490597.
@article{osti_1490597,
title = {Unraveling Microbial Communities Associated with Methylmercury Production in Paddy Soils},
author = {Liu, Yu-Rong and Johs, Alexander and Bi, Li and Lu, Xia and Hu, Hang-Wei and Sun, Dan and He, Ji-Zheng and Gu, Baohua},
abstractNote = {Here, we report rice consumption is now recognized as an important pathway of human exposure to the neurotoxin methylmercury (MeHg), particularly in countries where rice is a staple food. Although the discovery of a two-gene cluster hgcAB has linked Hg methylation to several phylogenetically diverse groups of anaerobic microorganisms converting inorganic mercury (Hg) to MeHg, the prevalence and diversity of Hg methylators in microbial communities of rice paddy soils remain unclear. We characterized the abundance and distribution of hgcAB genes using third-generation PacBio long-read sequencing and Illumina short-read metagenomic sequencing, in combination with quantitative PCR analyses in several mine-impacted paddy soils from southwest China. Both Illumina and PacBio sequencing analyses revealed that Hg methylating communities were dominated by iron-reducing bacteria (i.e., Geobacter) and methanogens, with a relatively low abundance of hgcA+ sulfate-reducing bacteria in the soil. A positive correlation was observed between the MeHg content in soil and the relative abundance of Geobacter carrying the hgcA gene. Phylogenetic analysis also uncovered some hgcAB sequences closely related to three novel Hg methylators, Geobacter anodireducens, Desulfuromonas sp. DDH964, and Desulfovibrio sp. J2, among which G. anodireducens was validated for its ability to methylate Hg. Lastly, these findings shed new light on microbial community composition and major clades likely driving Hg methylation in rice paddy soils.},
doi = {10.1021/acs.est.8b03052},
journal = {Environmental Science and Technology},
number = 22,
volume = 52,
place = {United States},
year = {Wed Oct 17 00:00:00 EDT 2018},
month = {Wed Oct 17 00:00:00 EDT 2018}
}
Web of Science
Figures / Tables:
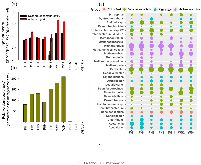 Fig. 1: Abundances and distribution of hgc$A$ genes from metagenomes for the selected paddy and upland soils obtained from Fenghuang (FH) and Wanshan (WS) mining areas in Southwest China; (a) Log10-transformed qPCR data for hgc$A$ gene copy number for Deltaproteobacteria and Archaea in the selected paddy soils, while qPCR formore »s yielded no hgc$A$. (b) Relative abundance ofhgc$A$ genes (from shotgun metagenomic sequencing) normalized to metagenome size (log Gbp); (c) Assignments of hgc$A$ genes from shotgun metagenomic sequencing at the genus level. Circle size represents the relative abundance of hgc$A$sequences assigned to a specific genus in each sample. Samples FHA-U, FHB-U and WSA-U were from upland fields.
Fig. 1: Abundances and distribution of hgc$A$ genes from metagenomes for the selected paddy and upland soils obtained from Fenghuang (FH) and Wanshan (WS) mining areas in Southwest China; (a) Log10-transformed qPCR data for hgc$A$ gene copy number for Deltaproteobacteria and Archaea in the selected paddy soils, while qPCR formore »s yielded no hgc$A$. (b) Relative abundance ofhgc$A$ genes (from shotgun metagenomic sequencing) normalized to metagenome size (log Gbp); (c) Assignments of hgc$A$ genes from shotgun metagenomic sequencing at the genus level. Circle size represents the relative abundance of hgc$A$sequences assigned to a specific genus in each sample. Samples FHA-U, FHB-U and WSA-U were from upland fields.
Works referenced in this record:
Methanogens: Principal Methylators of Mercury in Lake Periphyton
journal, September 2011
- Hamelin, Stéphanie; Amyot, Marc; Barkay, Tamar
- Environmental Science & Technology, Vol. 45, Issue 18
An introduction to geocatalysis
journal, June 1998
- Schoonen, Martin A. A.; Xu, Yong; Strongin, Daniel R.
- Journal of Geochemical Exploration, Vol. 62, Issue 1-3
Development and Validation of Broad-Range Qualitative and Clade-Specific Quantitative Molecular Probes for Assessing Mercury Methylation in the Environment
journal, July 2016
- Christensen, Geoff A.; Wymore, Ann M.; King, Andrew J.
- Applied and Environmental Microbiology, Vol. 82, Issue 19
Mercury as a Global Pollutant: Sources, Pathways, and Effects
journal, May 2013
- Driscoll, Charles T.; Mason, Robert P.; Chan, Hing Man
- Environmental Science & Technology, Vol. 47, Issue 10
Clustal W and Clustal X version 2.0
journal, September 2007
- Larkin, M. A.; Blackshields, G.; Brown, N. P.
- Bioinformatics, Vol. 23, Issue 21, p. 2947-2948
Human Exposure To Methylmercury through Rice Intake in Mercury Mining Areas, Guizhou Province, China
journal, January 2008
- Feng, Xinbin; Li, Ping; Qiu, Guangle
- Environmental Science & Technology, Vol. 42, Issue 1
Biological Methylation of Mercury in Aquatic Organisms
journal, August 1969
- Jensen, S.; JernelÖV, A.
- Nature, Vol. 223, Issue 5207
Cysteine Inhibits Mercury Methylation by Geobacter sulfurreducens PCA Mutant Δ omcBESTZ
journal, April 2015
- Lin, Hui; Lu, Xia; Liang, Liyuan
- Environmental Science & Technology Letters, Vol. 2, Issue 5
The Process of Methylmercury Accumulation in Rice ( Oryza sativa L.)
journal, April 2011
- Meng, Bo; Feng, Xinbin; Qiu, Guangle
- Environmental Science & Technology, Vol. 45, Issue 7
Mercury Methylation by Novel Microorganisms from New Environments
journal, September 2013
- Gilmour, Cynthia C.; Podar, Mircea; Bullock, Allyson L.
- Environmental Science & Technology, Vol. 47, Issue 20
Role of Settling Particles on Mercury Methylation in the Oxic Water Column of Freshwater Systems
journal, October 2016
- Gascón Díez, Elena; Loizeau, Jean-Luc; Cosio, Claudia
- Environmental Science & Technology, Vol. 50, Issue 21
Detection of a key Hg methylation gene, hgcA , in wetland soils : Detection of the Hg methylation gene, hgcA, in soils
journal, January 2014
- Schaefer, J. K.; Kronberg, R. -M.; Morel, F. M. M.
- Environmental Microbiology Reports, Vol. 6, Issue 5
CD-HIT: accelerated for clustering the next-generation sequencing data
journal, October 2012
- Fu, Limin; Niu, Beifang; Zhu, Zhengwei
- Bioinformatics, Vol. 28, Issue 23
Site-Directed Mutagenesis of HgcA and HgcB Reveals Amino Acid Residues Important for Mercury Methylation
journal, February 2015
- Smith, Steven D.; Bridou, Romain; Johs, Alexander
- Applied and Environmental Microbiology, Vol. 81, Issue 9
Environmental contamination in Antarctic ecosystems
journal, August 2008
- Bargagli, R.
- Science of The Total Environment, Vol. 400, Issue 1-3
Relationships between bacterial energetic metabolism, mercury methylation potential, and hgcA/hgcB gene expression in Desulfovibrio dechloroacetivorans BerOc1
journal, March 2015
- Goñi-Urriza, Marisol; Corsellis, Yannick; Lanceleur, Laurent
- Environmental Science and Pollution Research, Vol. 22, Issue 18
The Genetic Basis for Bacterial Mercury Methylation
journal, February 2013
- Parks, J. M.; Johs, A.; Podar, M.
- Science, Vol. 339, Issue 6125, p. 1332-1335
Global prevalence and distribution of genes and microorganisms involved in mercury methylation
journal, October 2015
- Podar, Mircea; Gilmour, Cynthia C.; Brandt, Craig C.
- Science Advances, Vol. 1, Issue 9
Mercury Methylation and Demethylation in Anoxic Lake Sediments and by Strictly Anaerobic Bacteria
journal, March 1998
- Pak, K. -R.; Bartha, R.
- Applied and Environmental Microbiology, Vol. 64, Issue 3
Syntrophic pathways for microbial mercury methylation
journal, March 2018
- Yu, Ri-Qing; Reinfelder, John R.; Hines, Mark E.
- The ISME Journal, Vol. 12, Issue 7
Rice methylmercury exposure and mitigation: A comprehensive review
journal, August 2014
- Rothenberg, Sarah E.; Windham-Myers, Lisamarie; Creswell, Joel E.
- Environmental Research, Vol. 133
Mercury Methylation by Dissimilatory Iron-Reducing Bacteria
journal, October 2006
- Kerin, E. J.; Gilmour, C. C.; Roden, E.
- Applied and Environmental Microbiology, Vol. 72, Issue 12, p. 7919-7921
Bioaccumulation of Methylmercury versus Inorganic Mercury in Rice ( Oryza sativa L.) Grain
journal, June 2010
- Zhang, Hua; Feng, Xinbin; Larssen, Thorjørn
- Environmental Science & Technology, Vol. 44, Issue 12
Cracking the Mercury Methylation Code
journal, March 2013
- Poulain, A. J.; Barkay, T.
- Science, Vol. 339, Issue 6125
Artifact formation of methyl mercury during aqueous distillation and alternative techniques for the extraction of methyl mercury from environmental samples
journal, June 1997
- Bloom, N. S.; Colman, John A.; Barber, Lee
- Fresenius' Journal of Analytical Chemistry, Vol. 358, Issue 3
Persistent Hg contamination and occurrence of Hg-methylating transcript (hgcA) downstream of a chlor-alkali plant in the Olt River (Romania)
journal, December 2015
- Bravo, Andrea G.; Loizeau, Jean-Luc; Dranguet, Perrine
- Environmental Science and Pollution Research, Vol. 23, Issue 11
Mercury Methylation from Unexpected Sources: Molybdate-Inhibited Freshwater Sediments and an Iron-Reducing Bacterium
journal, January 2006
- Fleming, E. J.; Mack, E. E.; Green, P. G.
- Applied and Environmental Microbiology, Vol. 72, Issue 1
Accelerated Profile HMM Searches
journal, October 2011
- Eddy, Sean R.
- PLoS Computational Biology, Vol. 7, Issue 10
Mercury Methylation by the Methanogen Methanospirillum hungatei
journal, August 2013
- Yu, Ri-Qing; Reinfelder, John R.; Hines, Mark E.
- Applied and Environmental Microbiology, Vol. 79, Issue 20
G eobacter sp. SD-1 with enhanced electrochemical activity in high-salt concentration solutions: Geobacter sp. SD-1 in high-salt solutions
journal, July 2014
- Sun, Dan; Call, Douglas; Wang, Aijie
- Environmental Microbiology Reports, Vol. 6, Issue 6
Anaerobic Mercury Methylation and Demethylation by Geobacter bemidjiensis Bem
journal, April 2016
- Lu, Xia; Liu, Yurong; Johs, Alexander
- Environmental Science & Technology, Vol. 50, Issue 8
Analysis of the Microbial Community Structure by Monitoring an Hg Methylation Gene (hgcA) in Paddy Soils along an Hg Gradient
journal, February 2014
- Liu, Y. -R.; Yu, R. -Q.; Zheng, Y. -M.
- Applied and Environmental Microbiology, Vol. 80, Issue 9
Ecological Patterns of nifH Genes in Four Terrestrial Climatic Zones Explored with Targeted Metagenomics Using FrameBot, a New Informatics Tool
journal, September 2013
- Wang, Qiong; Quensen, John F.; Fish, Jordan A.
- mBio, Vol. 4, Issue 5
Carbon Amendments Alter Microbial Community Structure and Net Mercury Methylation Potential in Sediments
journal, November 2017
- Christensen, Geoff A.; Somenahally, Anil C.; Moberly, James G.
- Applied and Environmental Microbiology, Vol. 84, Issue 3
Basic local alignment search tool
journal, October 1990
- Altschul, Stephen F.; Gish, Warren; Miller, Webb
- Journal of Molecular Biology, Vol. 215, Issue 3, p. 403-410
Long-Term Feeding of Elemental Sulfur Alters Microbial Community Structure and Eliminates Mercury Methylation Potential in Sulfate-Reducing Bacteria Abundant Activated Sludge
journal, March 2018
- Wang, Jin-ting; Zhang, Liang; Kang, Yuan
- Environmental Science & Technology, Vol. 52, Issue 8
Microbial mercury methylation in Antarctic sea ice
journal, August 2016
- Gionfriddo, Caitlin M.; Tate, Michael T.; Wick, Ryan R.
- Nature Microbiology, Vol. 1, Issue 10
Linking Microbial Activities and Low-Molecular-Weight Thiols to Hg Methylation in Biofilms and Periphyton from High-Altitude Tropical Lakes in the Bolivian Altiplano
journal, July 2018
- Bouchet, Sylvain; Goñi-Urriza, Marisol; Monperrus, Mathilde
- Environmental Science & Technology, Vol. 52, Issue 17
PacBio Sequencing and Its Applications
journal, October 2015
- Rhoads, Anthony; Au, Kin Fai
- Genomics, Proteomics & Bioinformatics, Vol. 13, Issue 5
Genome Sequence of the Piezophilic, Mesophilic Sulfate-Reducing Bacterium Desulfovibrio indicus J2 T
journal, April 2016
- Cao, Junwei; Maignien, Lois; Shao, Zongze
- Genome Announcements, Vol. 4, Issue 2
Depth profile of sulfate-reducing bacterial ribosomal RNA and mercury methylation in an estuarine sediment
journal, May 1996
- Devereux, Richard; Winfrey, Michael R.; Winfrey, Janet
- FEMS Microbiology Ecology, Vol. 20, Issue 1
Mercury flow through an Asian rice-based food web
journal, October 2017
- Abeysinghe, Kasun S.; Qiu, Guangle; Goodale, Eben
- Environmental Pollution, Vol. 229
Geobacteraceae are important members of mercury-methylating microbial communities of sediments impacted by waste water releases
journal, January 2018
- Bravo, Andrea G.; Zopfi, Jakob; Buck, Moritz
- The ISME Journal, Vol. 12, Issue 3
Mercury methylation in rice paddies and its possible controlling factors in the Hg mining area, Guizhou province, Southwest China
journal, August 2016
- Zhao, Lei; Qiu, Guangle; Anderson, Christopher W. N.
- Environmental Pollution, Vol. 215
Isolation and Genomic Characterization of Desulfuromonas soudanensis WTL, a Metal- and Electrode-Reducing Bacterium from Anoxic Deep Subsurface Brine
journal, April 2016
- Badalamenti, Jonathan P.; Summers, Zarath M.; Chan, Chi Ho
- Frontiers in Microbiology
Works referencing / citing this record:
Biotic formation of methylmercury: A bio–physico–chemical conundrum
journal, November 2019
- Bravo, Andrea G.; Cosio, Claudia
- Limnology and Oceanography, Vol. 65, Issue 5
Intestinal Methylation and Demethylation of Mercury
journal, December 2018
- Li, Hong; Lin, Xiaoying; Zhao, Jiating
- Bulletin of Environmental Contamination and Toxicology, Vol. 102, Issue 5
An Improved hgcAB Primer Set and Direct High-Throughput Sequencing Expand Hg-Methylator Diversity in Nature
journal, October 2020
- Gionfriddo, Caitlin M.; Wymore, Ann M.; Jones, Daniel S.
- Frontiers in Microbiology, Vol. 11
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal