In Silico Identification of Microbial Partners to Form Consortia with Anaerobic Fungi
Abstract
Lignocellulose is an abundant and renewable resource that holds great promise for sustainable bioprocessing. However, unpretreated lignocellulose is recalcitrant to direct utilization by most microbes. Current methods to overcome this barrier include expensive pretreatment steps to liberate cellulose and hemicellulose from lignin. Anaerobic gut fungi possess complex cellulolytic machinery specifically evolved to decompose crude lignocellulose, but they are not yet genetically tractable and have not been employed in industrial bioprocesses. Here, we aim to exploit the biomass-degrading abilities of anaerobic fungi by pairing them with another organism that can convert the fermentable sugars generated from hydrolysis into bioproducts. By combining experiments measuring the amount of excess fermentable sugars released by the fungal enzymes acting on crude lignocellulose, and a novel dynamic flux balance analysis algorithm, we screened potential consortia partners by qualitative suitability. Microbial growth simulations reveal that the fungus Anaeromyces robustus is most suited to pair with either the bacterium Clostridia ljungdahlii or the methanogen Methanosarcina barkeri—both organisms also found in the rumen microbiome. By capitalizing on simulations to screen six alternative organisms, valuable experimental time is saved towards identifying stable consortium members. This approach is also readily generalizable to larger systems and allows one to rationally select partnermore »
- Authors:
-
- Univ. of California, Santa Barbara, CA (United States). Department of Chemical Engineering
- Univ. of California, Santa Barbara, CA (United States). Department of Computer Science
- Publication Date:
- Research Org.:
- Univ. of California, Santa Barbara, CA (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC), Biological and Environmental Research (BER). Biological Systems Science Division
- OSTI Identifier:
- 1485152
- Grant/Contract Number:
- SC0010352
- Resource Type:
- Accepted Manuscript
- Journal Name:
- Processes
- Additional Journal Information:
- Journal Volume: 6; Journal Issue: 1; Journal ID: ISSN 2227-9717
- Publisher:
- Multidisciplinary Digital Publishing Institute (MDPI)
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES; anaerobic fungi; in silico modeling; microbial consortia; dynamic flux balance analysis; non-model organism; lignocellulose
Citation Formats
Wilken, St., Saxena, Mohan, Petzold, Linda, and O’Malley, Michelle. In Silico Identification of Microbial Partners to Form Consortia with Anaerobic Fungi. United States: N. p., 2018.
Web. doi:10.3390/pr6010007.
Wilken, St., Saxena, Mohan, Petzold, Linda, & O’Malley, Michelle. In Silico Identification of Microbial Partners to Form Consortia with Anaerobic Fungi. United States. https://doi.org/10.3390/pr6010007
Wilken, St., Saxena, Mohan, Petzold, Linda, and O’Malley, Michelle. Mon .
"In Silico Identification of Microbial Partners to Form Consortia with Anaerobic Fungi". United States. https://doi.org/10.3390/pr6010007. https://www.osti.gov/servlets/purl/1485152.
@article{osti_1485152,
title = {In Silico Identification of Microbial Partners to Form Consortia with Anaerobic Fungi},
author = {Wilken, St. and Saxena, Mohan and Petzold, Linda and O’Malley, Michelle},
abstractNote = {Lignocellulose is an abundant and renewable resource that holds great promise for sustainable bioprocessing. However, unpretreated lignocellulose is recalcitrant to direct utilization by most microbes. Current methods to overcome this barrier include expensive pretreatment steps to liberate cellulose and hemicellulose from lignin. Anaerobic gut fungi possess complex cellulolytic machinery specifically evolved to decompose crude lignocellulose, but they are not yet genetically tractable and have not been employed in industrial bioprocesses. Here, we aim to exploit the biomass-degrading abilities of anaerobic fungi by pairing them with another organism that can convert the fermentable sugars generated from hydrolysis into bioproducts. By combining experiments measuring the amount of excess fermentable sugars released by the fungal enzymes acting on crude lignocellulose, and a novel dynamic flux balance analysis algorithm, we screened potential consortia partners by qualitative suitability. Microbial growth simulations reveal that the fungus Anaeromyces robustus is most suited to pair with either the bacterium Clostridia ljungdahlii or the methanogen Methanosarcina barkeri—both organisms also found in the rumen microbiome. By capitalizing on simulations to screen six alternative organisms, valuable experimental time is saved towards identifying stable consortium members. This approach is also readily generalizable to larger systems and allows one to rationally select partner microbes for formation of stable consortia with non-model microbes like anaerobic fungi.},
doi = {10.3390/pr6010007},
journal = {Processes},
number = 1,
volume = 6,
place = {United States},
year = {Mon Jan 15 00:00:00 EST 2018},
month = {Mon Jan 15 00:00:00 EST 2018}
}
Web of Science
Figures / Tables:
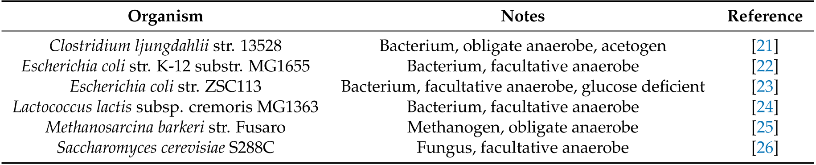 Table 1: Genome-scale models of potential consortia partners for the un-modeled anaerobic gut fungi used in this work.
Table 1: Genome-scale models of potential consortia partners for the un-modeled anaerobic gut fungi used in this work.
Works referenced in this record:
Genetic manipulation of Methanosarcina spp.
journal, January 2012
- Kohler, Petra R. A.; Metcalf, William W.
- Frontiers in Microbiology, Vol. 3
Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction
journal, January 2013
- Resch, Michael G.; Donohoe, Bryon S.; Baker, John O.
- Energy & Environmental Science, Vol. 6, Issue 6
BiGG Models: A platform for integrating, standardizing and sharing genome-scale models
journal, October 2015
- King, Zachary A.; Lu, Justin; Dräger, Andreas
- Nucleic Acids Research, Vol. 44, Issue D1
Connecting extracellular metabolomic measurements to intracellular flux states in yeast
journal, January 2009
- Mo, Monica L.; Palsson, Bernhard Ø; Herrgård, Markus J.
- BMC Systems Biology, Vol. 3, Issue 1
A review of genome-scale metabolic flux modeling of anaerobiosis in biotechnology
journal, November 2014
- Senger, Ryan S.; Yen, Jiun Y.; Fong, Stephen S.
- Current Opinion in Chemical Engineering, Vol. 6
Dynamic flux balance analysis for synthetic microbial communities
journal, October 2014
- Henson, Michael A.; Hanly, Timothy J.
- IET Systems Biology, Vol. 8, Issue 5
Co-culture systems and technologies: taking synthetic biology to the next level
journal, July 2014
- Goers, Lisa; Freemont, Paul; Polizzi, Karen M.
- Journal of The Royal Society Interface, Vol. 11, Issue 96
Biological pretreatment of lignocellulosic biomass – An overview
journal, January 2016
- Sindhu, Raveendran; Binod, Parameswaran; Pandey, Ashok
- Bioresource Technology, Vol. 199
Determination of growth of anaerobic fungi on soluble and cellulosic substrates using a pressure transducer
journal, March 1995
- Theodorou, M. K.; Davies, D. R.; Nielsen, B. B.
- Microbiology, Vol. 141, Issue 3
Comparison of growth characteristics of anaerobic fungi isolated from ruminant and non-ruminant herbivores during cultivation in a defined medium
journal, June 1991
- Teunissen, M. J.; Op den Camp, H. J. M.; Orpin, C. G.
- Journal of General Microbiology, Vol. 137, Issue 6
Unstructured Modeling of a Synthetic Microbial Consortium for Consolidated Production of Ethanol
journal, January 2013
- Hanly, Timothy J.; Henson, Michael A.
- IFAC Proceedings Volumes, Vol. 46, Issue 31
Anaerobic fungi (phylum Neocallimastigomycota ): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential
journal, August 2014
- Gruninger, Robert J.; Puniya, Anil K.; Callaghan, Tony M.
- FEMS Microbiology Ecology, Vol. 90, Issue 1
Consolidated bioprocessing of cellulosic biomass: an update
journal, October 2005
- Lynd, Lee R.; van Zyl, Willem H.; McBride, John E.
- Current Opinion in Biotechnology, Vol. 16, Issue 5, p. 577-583
Engineering microbial consortia: a new frontier in synthetic biology
journal, September 2008
- Brenner, Katie; You, Lingchong; Arnold, Frances H.
- Trends in Biotechnology, Vol. 26, Issue 9, p. 483-489
Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: concepts and recent developments
journal, August 2014
- Saini, Jitendra Kumar; Saini, Reetu; Tewari, Lakshmi
- 3 Biotech, Vol. 5, Issue 4
The importance of sourcing enzymes from non-conventional fungi for metabolic engineering and biomass breakdown
journal, November 2017
- Seppälä, Susanna; Wilken, St. Elmo; Knop, Doriv
- Metabolic Engineering, Vol. 44
Dynamic flux balance modeling of S. cerevisiae and E. coli co-cultures for efficient consumption of glucose/xylose mixtures
journal, October 2011
- Hanly, Timothy J.; Urello, Morgan; Henson, Michael A.
- Applied Microbiology and Biotechnology, Vol. 93, Issue 6
The effects of alternate optimal solutions in constraint-based genome-scale metabolic models
journal, October 2003
- Mahadevan, R.; Schilling, C. H.
- Metabolic Engineering, Vol. 5, Issue 4
Fuelling the future: microbial engineering for the production of sustainable biofuels
journal, March 2016
- Liao, James C.; Mi, Luo; Pontrelli, Sammy
- Nature Reviews Microbiology, Vol. 14, Issue 5
Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli
journal, November 2011
- Bokinsky, G.; Peralta-Yahya, P. P.; George, A.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 50, p. 19949-19954
Construction of robust dynamic genome-scale metabolic model structures of Saccharomyces cerevisiae through iterative re-parameterization
journal, September 2014
- Sánchez, Benjamín J.; Pérez-Correa, José R.; Agosin, Eduardo
- Metabolic Engineering, Vol. 25
Genome-Based Modeling and Design of Metabolic Interactions in Microbial Communities
journal, October 2012
- Mahadevan, Radhakrishnan; Henson, Michael A.
- Computational and Structural Biotechnology Journal, Vol. 3, Issue 4
Modeling methanogenesis with a genome‐scale metabolic reconstruction of Methanosarcina barkeri
journal, January 2006
- Feist, Adam M.; Scholten, Johannes C. M.; Palsson, Bernhard Ø
- Molecular Systems Biology, Vol. 2, Issue 1
ll-ACHRB: a scalable algorithm for sampling the feasible solution space of metabolic networks
journal, March 2016
- Saa, Pedro A.; Nielsen, Lars K.
- Bioinformatics, Vol. 32, Issue 15
Steady-state and dynamic flux balance analysis of ethanol production by Saccharomyces cerevisiae
journal, May 2009
- Hjersted, J. L.; Henson, M. A.
- IET Systems Biology, Vol. 3, Issue 3
Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes
journal, February 2016
- Solomon, Kevin V.; Haitjema, Charles H.; Henske, John K.
- Science, Vol. 351, Issue 6278
Dynamic flux balance modeling of microbial co-cultures for efficient batch fermentation of glucose and xylose mixtures
journal, October 2010
- Hanly, Timothy J.; Henson, Michael A.
- Biotechnology and Bioengineering, Vol. 108, Issue 2
Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose
journal, January 2018
- Henske, John K.; Wilken, St. Elmo; Solomon, Kevin V.
- Biotechnology and Bioengineering, Vol. 115, Issue 4
DFBAlab: a fast and reliable MATLAB code for dynamic flux balance analysis
journal, December 2014
- Gomez, Jose A.; Höffner, Kai; Barton, Paul I.
- BMC Bioinformatics, Vol. 15, Issue 1
Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass
journal, August 2013
- Minty, Jeremy J.; Singer, Marc E.; Scholz, Scott A.
- Proceedings of the National Academy of Sciences, Vol. 110, Issue 36
Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii
journal, January 2013
- Nagarajan, Harish; Sahin, Merve; Nogales, Juan
- Microbial Cell Factories, Vol. 12, Issue 1
Genome-scale metabolic model for Lactococcus lactis MG1363 and its application to the analysis of flavor formation
journal, August 2013
- Flahaut, Nicolas A. L.; Wiersma, Anne; van de Bunt, Bert
- Applied Microbiology and Biotechnology, Vol. 97, Issue 19
Industrial systems biology
journal, February 2010
- Otero, José Manuel; Nielsen, Jens
- Biotechnology and Bioengineering, Vol. 105, Issue 3
Anaerobic gut fungi: Advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production: Anaerobic Gut Fungi
journal, May 2014
- Haitjema, Charles H.; Solomon, Kevin V.; Henske, John K.
- Biotechnology and Bioengineering, Vol. 111, Issue 8
Characterization of hydrogenosomes and their role in glucose metabolism of Neocallimastix sp. L2
journal, November 1993
- Marvin-Sikkema, FemkeD.; Pedro Gomes, TeresaM.; Grivet, Jean-Philippe
- Archives of Microbiology, Vol. 160, Issue 5
A co-fermentation strategy to consume sugar mixtures effectively
journal, January 2008
- Eiteman, Mark A.; Lee, Sarah A.; Altman, Elliot
- Journal of Biological Engineering, Vol. 2, Issue 1
Review Article: The hydrogenosome
journal, December 1993
- Muller, M.
- Journal of General Microbiology, Vol. 139, Issue 12
Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential?
journal, October 2009
- Alper, Hal; Stephanopoulos, Gregory
- Nature Reviews Microbiology, Vol. 7, Issue 10
Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina)
journal, May 2008
- Martinez, Diego; Berka, Randy M.; Henrissat, Bernard
- Nature Biotechnology, Vol. 26, Issue 5
iML1515, a knowledgebase that computes Escherichia coli traits
journal, October 2017
- Monk, Jonathan M.; Lloyd, Colton J.; Brunk, Elizabeth
- Nature Biotechnology, Vol. 35, Issue 10
What is flux balance analysis?
journal, March 2010
- Orth, Jeffrey D.; Thiele, Ines; Palsson, Bernhard Ø
- Nature Biotechnology, Vol. 28, Issue 3
A reliable simulator for dynamic flux balance analysis
journal, October 2012
- Höffner, K.; Harwood, S. M.; Barton, P. I.
- Biotechnology and Bioengineering, Vol. 110, Issue 3
Microbial communities for bioprocessing: lessons learned from nature
journal, November 2016
- Peng, Xuefeng “Nick”; Gilmore, Sean P.; O’Malley, Michelle A.
- Current Opinion in Chemical Engineering, Vol. 14
Phosphorylation of D-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase.
journal, January 1975
- Curtis, S. J.; Epstein, W.
- Journal of Bacteriology, Vol. 122, Issue 3
Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110.
journal, January 1994
- Varma, A.; Palsson, B. O.
- Applied and Environmental Microbiology, Vol. 60, Issue 10
Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose
journal, January 2018
- Henske, John K.; Wilken, St. Elmo; Solomon, Kevin V.
- Biotechnology and Bioengineering, Vol. 115, Issue 4
Genome-scale metabolic model for Lactococcus lactis MG1363 and its application to the analysis of flavor formation
journal, August 2013
- Flahaut, Nicolas A. L.; Wiersma, Anne; van de Bunt, Bert
- Applied Microbiology and Biotechnology, Vol. 97, Issue 19
Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: concepts and recent developments
journal, August 2014
- Saini, Jitendra Kumar; Saini, Reetu; Tewari, Lakshmi
- 3 Biotech, Vol. 5, Issue 4
Steady-state and dynamic flux balance analysis of ethanol production by Saccharomyces cerevisiae
journal, May 2009
- Hjersted, J. L.; Henson, M. A.
- IET Systems Biology, Vol. 3, Issue 3
A co-fermentation strategy to consume sugar mixtures effectively
journal, January 2008
- Eiteman, Mark A.; Lee, Sarah A.; Altman, Elliot
- Journal of Biological Engineering, Vol. 2, Issue 1
DFBAlab: a fast and reliable MATLAB code for dynamic flux balance analysis
journal, December 2014
- Gomez, Jose A.; Höffner, Kai; Barton, Paul I.
- BMC Bioinformatics, Vol. 15, Issue 1
Works referencing / citing this record:
Biomass-degrading enzymes are catabolite repressed in anaerobic gut fungi
journal, October 2018
- Henske, John K.; Gilmore, Sean P.; Haitjema, Charles H.
- AIChE Journal, Vol. 64, Issue 12
Special Issue: Microbial Community Modeling: Prediction of Microbial Interactions and Community Dynamics
journal, April 2018
- Song, Hyun-Seob
- Processes, Vol. 6, Issue 5
Predicting the Longitudinally and Radially Varying Gut Microbiota Composition Using Multi-Scale Microbial Metabolic Modeling
journal, June 2019
- Chan,
- Processes, Vol. 7, Issue 7
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal