Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae
Abstract
Planctomyces brasiliensis Schlesner 1990 belongs to the order Planctomycetales, which differs from other bacterial taxa by several distinctive features such as internal cell compartmentalization, multiplication by forming buds directly from the spherical, ovoid or pear-shaped mother cell and a cell wall consisting of a proteinaceous layer rather than a peptidoglycan layer. The first strains of P. brasiliensis, including the type strain IFAM 1448T, were isolated from a water sample of Lagoa Vermelha, a salt pit near Rio de Janeiro, Brasil. This is the second completed genome sequence of a type strain of the genus Planctomyces to be published and the sixth type strain genome sequence from the family Planctomycetaceae. The 6,006,602 bp long genome with its 4,811 protein-coding and 54 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project. Phylogenomic analyses indicate that the classification within the Planctomycetaceae is partially in conflict with its evolutionary history, as the positioning of Schlesneria renders the genus Planctomyces paraphyletic. A re-analysis of published fatty-acid measurements also does not support the current arrangement of the two genera. A quantitative comparison of phylogenetic and phenotypic aspects indicates that the three Planctomyces species with type strains available in public culturemore »
- Authors:
-
more »
- DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig (Germany)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States); Los Alamos National Lab. (LANL), Los Alamos, NM (United States)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States) ; Los Alamos National Lab. (LANL), Los Alamos, NM (United States)
- Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States) ; Oak Ridge National Lab. (ORNL), Oak Ridge, TN (United States)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States) ; Oak Ridge National Lab. (ORNL), Oak Ridge, TN (United States)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States); Oak Ridge National Lab. (ORNL), Oak Ridge, TN (United States)
- Jomo Kenyatta University of Agriculture and Technology, Nairobi (Kenya)
- Helmholtz Centre for Infection Research (HZI), Braunschweig (Germany)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States); Univ. of California, Davis, CA (United States)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States); Univ. of Queensland, Brisbane (Australia)
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States); King Abdulaziz Univ., Jeddah (Saudi Arabia)
- Publication Date:
- Research Org.:
- Oak Ridge National Laboratory (ORNL), Oak Ridge, TN (United States); Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States)
- Sponsoring Org.:
- Work for Others (WFO); USDOE Office of Science (SC), Biological and Environmental Research (BER)
- OSTI Identifier:
- 1286834
- Alternate Identifier(s):
- OSTI ID: 1511412
- Grant/Contract Number:
- AC05-00OR22725; AC02-05CH11231
- Resource Type:
- Accepted Manuscript
- Journal Name:
- Standards in Genomic Sciences
- Additional Journal Information:
- Journal Volume: 9; Journal Issue: 10; Journal ID: ISSN 1944-3277
- Publisher:
- BioMed Central
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES; Non-peptidoglycan bacteria; Stalked bacteria; Halotolerant; Gram-negative; Taxonomic descriptions; Planctomycetales; Planctomycetes; GEBA
Citation Formats
Scheuner, Carmen, Tindall, Brian J., Lu, Megan, Nolan, Matt, Lapidus, Alla L., Cheng, Jan-Fang, Goodwin, Lynne A., Pitluck, Sam, Huntemann, Marcel, Liolios, Konstantinos, Pagani, Ioanna, Mavromatis, Konstantinos, Ivanova, Natalia, Pati, Amrita, Chen, Amy, Palaniappan, Krishna, Jeffries, Cynthia D., Hauser, Loren John, Land, Miriam L., Mwirichia, Romano, Rohde, Manfred, Abt, Birte, Detter, John Chris, Woyke, Tanja, Eisen, Jonathan A., Markowitz, Victor, Hugenholtz, Philip, Goker, Markus, Kyrpides, Nikos C., and Klenk, Hans-Peter. Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. United States: N. p., 2014.
Web. doi:10.1186/1944-3277-9-10.
Scheuner, Carmen, Tindall, Brian J., Lu, Megan, Nolan, Matt, Lapidus, Alla L., Cheng, Jan-Fang, Goodwin, Lynne A., Pitluck, Sam, Huntemann, Marcel, Liolios, Konstantinos, Pagani, Ioanna, Mavromatis, Konstantinos, Ivanova, Natalia, Pati, Amrita, Chen, Amy, Palaniappan, Krishna, Jeffries, Cynthia D., Hauser, Loren John, Land, Miriam L., Mwirichia, Romano, Rohde, Manfred, Abt, Birte, Detter, John Chris, Woyke, Tanja, Eisen, Jonathan A., Markowitz, Victor, Hugenholtz, Philip, Goker, Markus, Kyrpides, Nikos C., & Klenk, Hans-Peter. Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. United States. https://doi.org/10.1186/1944-3277-9-10
Scheuner, Carmen, Tindall, Brian J., Lu, Megan, Nolan, Matt, Lapidus, Alla L., Cheng, Jan-Fang, Goodwin, Lynne A., Pitluck, Sam, Huntemann, Marcel, Liolios, Konstantinos, Pagani, Ioanna, Mavromatis, Konstantinos, Ivanova, Natalia, Pati, Amrita, Chen, Amy, Palaniappan, Krishna, Jeffries, Cynthia D., Hauser, Loren John, Land, Miriam L., Mwirichia, Romano, Rohde, Manfred, Abt, Birte, Detter, John Chris, Woyke, Tanja, Eisen, Jonathan A., Markowitz, Victor, Hugenholtz, Philip, Goker, Markus, Kyrpides, Nikos C., and Klenk, Hans-Peter. Mon .
"Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae". United States. https://doi.org/10.1186/1944-3277-9-10. https://www.osti.gov/servlets/purl/1286834.
@article{osti_1286834,
title = {Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae},
author = {Scheuner, Carmen and Tindall, Brian J. and Lu, Megan and Nolan, Matt and Lapidus, Alla L. and Cheng, Jan-Fang and Goodwin, Lynne A. and Pitluck, Sam and Huntemann, Marcel and Liolios, Konstantinos and Pagani, Ioanna and Mavromatis, Konstantinos and Ivanova, Natalia and Pati, Amrita and Chen, Amy and Palaniappan, Krishna and Jeffries, Cynthia D. and Hauser, Loren John and Land, Miriam L. and Mwirichia, Romano and Rohde, Manfred and Abt, Birte and Detter, John Chris and Woyke, Tanja and Eisen, Jonathan A. and Markowitz, Victor and Hugenholtz, Philip and Goker, Markus and Kyrpides, Nikos C. and Klenk, Hans-Peter},
abstractNote = {Planctomyces brasiliensis Schlesner 1990 belongs to the order Planctomycetales, which differs from other bacterial taxa by several distinctive features such as internal cell compartmentalization, multiplication by forming buds directly from the spherical, ovoid or pear-shaped mother cell and a cell wall consisting of a proteinaceous layer rather than a peptidoglycan layer. The first strains of P. brasiliensis, including the type strain IFAM 1448T, were isolated from a water sample of Lagoa Vermelha, a salt pit near Rio de Janeiro, Brasil. This is the second completed genome sequence of a type strain of the genus Planctomyces to be published and the sixth type strain genome sequence from the family Planctomycetaceae. The 6,006,602 bp long genome with its 4,811 protein-coding and 54 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project. Phylogenomic analyses indicate that the classification within the Planctomycetaceae is partially in conflict with its evolutionary history, as the positioning of Schlesneria renders the genus Planctomyces paraphyletic. A re-analysis of published fatty-acid measurements also does not support the current arrangement of the two genera. A quantitative comparison of phylogenetic and phenotypic aspects indicates that the three Planctomyces species with type strains available in public culture collections should be placed in separate genera. Thus the genera Gimesia, Planctopirus and Rubinisphaera are proposed to accommodate P. maris, P. limnophilus and P. brasiliensis, respectively. Pronounced differences between the reported G + C content of Gemmata obscuriglobus, Singulisphaera acidiphila and Zavarzinella formosa and G + C content calculated from their genome sequences call for emendation of their species descriptions. In addition to other features, the range of G + C values reported for the genera within the Planctomycetaceae indicates that the descriptions of the family and the order should be emended.},
doi = {10.1186/1944-3277-9-10},
journal = {Standards in Genomic Sciences},
number = 10,
volume = 9,
place = {United States},
year = {Mon Dec 08 00:00:00 EST 2014},
month = {Mon Dec 08 00:00:00 EST 2014}
}
Web of Science
Figures / Tables:
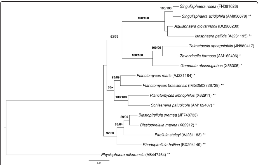 Figure 1: Phylogenetic tree highlighting the position of P. brasiliensis relative to the other species within the family Planctomycetaceae. The tree was inferred from 1,343 aligned characters of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion as previously described. Rooting was done initially using the midpoint methodmore »
Figure 1: Phylogenetic tree highlighting the position of P. brasiliensis relative to the other species within the family Planctomycetaceae. The tree was inferred from 1,343 aligned characters of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion as previously described. Rooting was done initially using the midpoint methodmore »
Works referenced in this record:
Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a Siberian peat bog
journal, February 2009
- Kulichevskaya, I. S.; Baulina, O. I.; Bodelier, P. L. E.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 59, Issue 2
Parsimony, likelihood, and simplicity
journal, April 2003
- Goloboff, Pablo A.
- Cladistics, Vol. 19, Issue 2
The PVC superphylum: exceptions to the bacterial definition?
journal, August 2013
- Fuerst, John A.
- Antonie van Leeuwenhoek, Vol. 104, Issue 4
Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes
journal, January 2009
- Fukunaga, Yukiyo; Kurahashi, Midori; Sakiyama, Yayoi
- The Journal of General and Applied Microbiology, Vol. 55, Issue 4
Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis
journal, April 2000
- Castresana, J.
- Molecular Biology and Evolution, Vol. 17, Issue 4
Phylogenetic Systematics
journal, January 1965
- Hennig, Willi
- Annual Review of Entomology, Vol. 10, Issue 1
Approved Lists of Bacterial Names
journal, January 1980
- Sneath, P. H. A.; McGOWAN, Vicki; Skerman, V. B. D.
- International Journal of Systematic and Evolutionary Microbiology, Vol. 30, Issue 1
A Phylogenomic Approach to Resolve the Arthropod Tree of Life
journal, June 2010
- Meusemann, K.; von Reumont, B. M.; Simon, S.
- Molecular Biology and Evolution, Vol. 27, Issue 11
Planktonic microbial community composition across steep physical/chemical gradients in permanently ice-covered Lake Bonney, Antarctica
journal, March 2006
- Glatz, R. E.; Lepp, P. W.; Ward, B. B.
- Geobiology, Vol. 4, Issue 1
Gapped BLAST and PSI-BLAST: a new generation of protein database search programs
journal, September 1997
- Altschul, Stephen F.; Madden, Thomas L.; Schäffer, Alejandro A.
- Nucleic Acids Research, Vol. 25, Issue 17, p. 3389-3402
The Information Content of the Phylogenetic System
journal, December 1979
- Farris, James S.
- Systematic Zoology, Vol. 28, Issue 4
Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya.
journal, June 1990
- Woese, C. R.; Kandler, O.; Wheelis, M. L.
- Proceedings of the National Academy of Sciences, Vol. 87, Issue 12
Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov.
journal, October 1986
- Schlesner, Heinz; Stackebrandt, Erko
- Systematic and Applied Microbiology, Vol. 8, Issue 3
Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses
journal, October 2010
- Yarza, Pablo; Ludwig, Wolfgang; Euzéby, Jean
- Systematic and Applied Microbiology, Vol. 33, Issue 6
Genome sequence-based species delimitation with confidence intervals and improved distance functions
journal, January 2013
- Meier-Kolthoff, Jan P.; Auch, Alexander F.; Klenk, Hans-Peter
- BMC Bioinformatics, Vol. 14, Issue 1
Prodigal: prokaryotic gene recognition and translation initiation site identification
journal, March 2010
- Hyatt, Doug; Chen, Gwo-Liang; LoCascio, Philip F.
- BMC Bioinformatics, Vol. 11, Issue 1
Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell
journal, June 2001
- Lindsay, Margaret; Webb, Richard; Strous, Marc
- Archives of Microbiology, Vol. 175, Issue 6
Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age
journal, February 2014
- Klenk, Hans-Peter; Meier-Kolthoff, Jan P.; Göker, Markus
- International Journal of Systematic and Evolutionary Microbiology, Vol. 64, Issue 2
VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R
journal, January 2011
- Chen, Hanbo; Boutros, Paul C.
- BMC Bioinformatics, Vol. 12, Issue 1
Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands
journal, May 2008
- Kulichevskaya, I. S.; Ivanova, A. O.; Baulina, O. I.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 58, Issue 5
Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21T)
journal, October 2011
- Chang, Yun-juan; Land, Miriam; Hauser, Loren
- Standards in Genomic Sciences, Vol. 5, Issue 1
An empirical test of the midpoint rooting method: A TEST FOR THE MIDPOINT ROOTING METHOD
journal, December 2007
- Hess, Pablo N.; De Moraes Russo, Claudia A.
- Biological Journal of the Linnean Society, Vol. 92, Issue 4
Assignment of ATCC 27377 to Pirella gen. nov. as Pirella staleyi comb. nov.
journal, October 1984
- Schlesner, H.; Hirsch, P.
- International Journal of Systematic Bacteriology, Vol. 34, Issue 4
Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula
journal, September 2004
- Schlesner, H.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 54, Issue 5
Planctomyces brasiliensis sp. nov., a Halotolerant Bacterium from a Salt Pit
journal, October 1989
- Schlesner, Heinz
- Systematic and Applied Microbiology, Vol. 12, Issue 2
Complete genome sequence of the termite hindgut bacterium Spirochaeta coccoides type strain (SPN1T), reclassification in the genus Sphaerochaeta as Sphaerochaeta coccoides comb. nov. and emendations of the family Spirochaetaceae and the genus Sphaerochaeta
journal, May 2012
- Abt, Birte; Han, Cliff; Scheuner, Carmen
- Standards in Genomic Sciences, Vol. 6, Issue 2
Phylogeny-driven target selection for large-scale genome-sequencing (and other) projects
journal, May 2013
- Göker, Markus; Klenk, Hans-Peter
- Standards in Genomic Sciences, Vol. 8, Issue 2
Singulisphaera rosea sp. nov., a planctomycete from acidic Sphagnum peat, and emended description of the genus Singulisphaera
journal, February 2011
- Kulichevskaya, I. S.; Detkova, E. N.; Bodelier, P. L. E.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 62, Issue 1
Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin
journal, December 2013
- Devos, Damien P.
- Antonie van Leeuwenhoek, Vol. 105, Issue 2
Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs
journal, January 2010
- Auch, Alexander F.; Klenk, Hans-Peter; Göker, Markus
- Standards in Genomic Sciences, Vol. 2, Issue 1
Complete genome sequence of Planctomyces limnophilus type strain (Mü 290T)
journal, July 2010
- LaButti, Kurt; Sikorski, Johannes; Schneider, Susanne
- Standards in Genomic Sciences, Vol. 3, Issue 1
Genomic Evolution of 11 Type Strains within Family Planctomycetaceae
journal, January 2014
- Guo, Min; Zhou, Qian; Zhou, Yizhuang
- PLoS ONE, Vol. 9, Issue 1
A Rapid Bootstrap Algorithm for the RAxML Web Servers
journal, October 2008
- Stamatakis, Alexandros; Hoover, Paul; Rougemont, Jacques
- Systematic Biology, Vol. 57, Issue 5
The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata
journal, December 2011
- Pagani, I.; Liolios, K.; Jansson, J.
- Nucleic Acids Research, Vol. 40, Issue D1
Complete genome sequence of Pirellula staleyi type strain (ATCC 27377T)
journal, December 2009
- Clum, Alicia; Tindall, Brian J.; Sikorski, Johannes
- Standards in Genomic Sciences, Vol. 1, Issue 3
An efficient algorithm for large-scale detection of protein families
journal, April 2002
- Enright, A. J.
- Nucleic Acids Research, Vol. 30, Issue 7
MUSCLE: multiple sequence alignment with high accuracy and high throughput
journal, March 2004
- Edgar, R. C.
- Nucleic Acids Research, Vol. 32, Issue 5, p. 1792-1797
Phylogenetic diversity, polyamine pattern and DNA base composition of members of the order Planctomycetales
journal, April 1999
- Griepenburg, U.; Ward-Rainey, N.; Mohamed, S.
- International Journal of Systematic Bacteriology, Vol. 49, Issue 2
Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from Sphagnum-dominated boreal wetlands
journal, November 2007
- Kulichevskaya, I. S.; Ivanova, A. O.; Belova, S. E.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 57, Issue 11
The Fast Changing Landscape of Sequencing Technologies and Their Impact on Microbial Genome Assemblies and Annotation
journal, December 2012
- Mavromatis, Konstantinos; Land, Miriam L.; Brettin, Thomas S.
- PLoS ONE, Vol. 7, Issue 12
IMG ER: a system for microbial genome annotation expert review and curation
journal, June 2009
- Markowitz, Victor M.; Mavromatis, Konstantinos; Ivanova, Natalia N.
- Bioinformatics, Vol. 25, Issue 17
A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea
journal, December 2009
- Wu, Dongying; Hugenholtz, Philip; Mavromatis, Konstantinos
- Nature, Vol. 462, Issue 7276
Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group
journal, September 1986
- Liesack, W.; K�nig, H.; Schlesner, H.
- Archives of Microbiology, Vol. 145, Issue 4
Planctomyces stranskae (ex Wawrik 1952) sp. nov., nom. rev. and Planctomyces guttaeformis (ex Hortobagyi 1965) sp. nov., nom. rev.
journal, October 1984
- Starr, M. P.; Schmidt, J. M.
- International Journal of Systematic Bacteriology, Vol. 34, Issue 4
The DOE-JGI Standard Operating Procedure for the Annotations of Microbial Genomes
journal, July 2009
- Mavromatis, Konstantinos; Ivanova, Natalia N.; Chen, I-Min A.
- Standards in Genomic Sciences, Vol. 1, Issue 1
Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology
journal, December 1971
- Fitch, Walter M.
- Systematic Zoology, Vol. 20, Issue 4
The DNA Bank Network: The Start from a German Initiative
journal, March 2011
- Gemeinholzer, Birgit; Dröge, Gabriele; Zetzsche, Holger
- Biopreservation and Biobanking, Vol. 9, Issue 1
Characterization of Planctomyces limnophilus and Development of Genetic Tools for Its Manipulation Establish It as a Model Species for the Phylum Planctomycetes
journal, July 2011
- Jogler, Christian; Glöckner, Frank Oliver; Kolter, Roberto
- Applied and Environmental Microbiology, Vol. 77, Issue 16
Codivergence of Mycoviruses with Their Hosts
journal, July 2011
- Göker, Markus; Scheuner, Carmen; Klenk, Hans-Peter
- PLoS ONE, Vol. 6, Issue 7
From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules
journal, August 2013
- Jeske, Olga; Jogler, Mareike; Petersen, Jörn
- Antonie van Leeuwenhoek, Vol. 104, Issue 4
“Who Has First Observed Planctomyces”
journal, April 2005
- Langó, Zsuzsanna
- Acta Microbiologica et Immunologica Hungarica, Vol. 52, Issue 1
Novel Insights into the Diversity of Catabolic Metabolism from Ten Haloarchaeal Genomes
journal, May 2011
- Anderson, Iain; Scheuner, Carmen; Göker, Markus
- PLoS ONE, Vol. 6, Issue 5
Chemotaxonomic Investigation of Various Prosthecate and/or Budding Bacteria
journal, April 1993
- Sittig, Manuel; Schlesner, Heinz
- Systematic and Applied Microbiology, Vol. 16, Issue 1
Complete genome sequence of Isosphaera pallida type strain (IS1BT)
journal, February 2011
- Göker, Markus; Cleland, David; Saunders, Elizabeth
- Standards in Genomic Sciences, Vol. 4, Issue 1
Complete genome sequence of Kytococcus sedentarius type strain (541T)
journal, July 2009
- Sims, David; Brettin, Thomas; Detter, John C.
- Standards in Genomic Sciences, Vol. 1, Issue 1
Planctomyces limnophilus sp. nov., a Stalked and Budding Bacterium from Freshwater
journal, December 1985
- Hirsch, Peter; Müller, Michael
- Systematic and Applied Microbiology, Vol. 6, Issue 3
Gene Ontology: tool for the unification of biology
journal, May 2000
- Ashburner, Michael; Ball, Catherine A.; Blake, Judith A.
- Nature Genetics, Vol. 25, Issue 1
The Information Content of the Phylogenetic System
journal, December 1979
- Farris, J. S.
- Systematic Biology, Vol. 28, Issue 4
The Genomic Standards Consortium
journal, June 2011
- Field, Dawn; Amaral-Zettler, Linda; Cochrane, Guy
- PLoS Biology, Vol. 9, Issue 6
Phylogenetic Systematics
text, January 2000
- Basibuyuk, Hasan H.; Belshaw, Robert; Bardakcı, Fevzi
- Önder Matbaa
RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models
journal, August 2006
- Stamatakis, Alexandros
- Bioinformatics, Vol. 22, Issue 21
The budding bacteria, Pirellula and Planctomyces, with atypical 16S rRNA and absence of peptidoglycan, show eubacterial phospholipids and uniquely high proportions of long chain beta-hydroxy fatty acids in the lipopolysaccharide lipid A
journal, January 1988
- Kerger, B. D.; Mancuso, C. A.; Nichols, P. D.
- Archives of Microbiology, Vol. 149, Issue 3
OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes
journal, September 2003
- Li, L.
- Genome Research, Vol. 13, Issue 9
Velvet: Algorithms for de novo short read assembly using de Bruijn graphs
journal, February 2008
- Zerbino, D. R.; Birney, E.
- Genome Research, Vol. 18, Issue 5
Blastopirellula cremea sp. nov., isolated from a dead ark clam
journal, November 2012
- Lee, H. -W.; Roh, S. W.; Shin, N. -R.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 63, Issue Pt 6
opm: an R package for analysing OmniLog(R) phenotype microarray data
journal, June 2013
- Vaas, L. A. I.; Sikorski, J.; Hofner, B.
- Bioinformatics, Vol. 29, Issue 14
Planctomyces maris sp. nov.: a Marine Isolate of the Planctomyces-Blastocaulis Group of Budding Bacteria
journal, November 1976
- Bauld, J.; Staley, J. T.
- Journal of General Microbiology, Vol. 97, Issue 1
Genome-Scale Phylogeny and the Detection of Systematic Biases
journal, July 2004
- Phillips, Matthew J.; Delsuc, Frédéric; Penny, David
- Molecular Biology and Evolution, Vol. 21, Issue 7
Transposon Mutagenesis of Planctomyces limnophilus and Analysis of a pckA Mutant
collection, January 2012
- Schreier, Harold J.; Dejtisakdi, Wipawee; Escalante, Jose Ortega
- American Society for Microbiology
Evolutionary trees from DNA sequences: A maximum likelihood approach
journal, November 1981
- Felsenstein, Joseph
- Journal of Molecular Evolution, Vol. 17, Issue 6
Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs
journal, April 1987
- Giovannoni, S. J.; Schabtach, E.; Castenholz, R. W.
- Archives of Microbiology, Vol. 147, Issue 3
How Many Bootstrap Replicates Are Necessary?
journal, March 2010
- Pattengale, Nicholas D.; Alipour, Masoud; Bininda-Emonds, Olaf R. P.
- Journal of Computational Biology, Vol. 17, Issue 3
Planctomyces maris sp. nov., nom. rev.
journal, October 1980
- Bauld, J.; Staley, J. T.
- International Journal of Systematic Bacteriology, Vol. 30, Issue 4
How Many Bootstrap Replicates Are Necessary?
book, January 2009
- Pattengale, Nicholas D.; Alipour, Masoud; Bininda-Emonds, Olaf R. P.
- Lecture Notes in Computer Science
Aquisphaera giovannonii gen. nov., sp. nov., a planctomycete isolated from a freshwater aquarium
journal, January 2011
- Bondoso, J.; Albuquerque, L.; Nobre, M. F.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 61, Issue 12
The minimum information about a genome sequence (MIGS) specification
journal, May 2008
- Field, Dawn; Garrity, George; Gray, Tanya
- Nature Biotechnology, Vol. 26, Issue 5
RASCAL: rapid scanning and correction of multiple sequence alignments
journal, June 2003
- Thompson, J. D.; Thierry, J. C.; Poch, O.
- Bioinformatics, Vol. 19, Issue 9
Comparative analysis of ribonuclease P RNA of the planctomycetes
journal, July 2004
- Butler, M. K.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 54, Issue 4
Pirella marina sp. nov., a budding, peptidoglycan-less bacterium from brackish water
journal, October 1986
- Schlesner, Heinz
- Systematic and Applied Microbiology, Vol. 8, Issue 3
Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology
journal, December 1971
- Fitch, W. M.
- Systematic Biology, Vol. 20, Issue 4
Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison
journal, January 2010
- Auch, Alexander F.; von Jan, Mathias; Klenk, Hans-Peter
- Standards in Genomic Sciences, Vol. 2, Issue 1
En route to a genome-based classification of Archaea and Bacteria?
journal, June 2010
- Klenk, H. -P.; Göker, M.
- Systematic and Applied Microbiology, Vol. 33, Issue 4
Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon
journal, January 1997
- Fuerst, J. A.; Gwilliam, H. G.; Lindsay, M.
- Applied and Environmental Microbiology, Vol. 63, Issue 1
Complete genome sequence of the thermophilic, hydrogen-oxidizing Bacillus tusciae type strain (T2T) and reclassification in the new genus, Kyrpidia gen. nov. as Kyrpidia tusciae comb. nov. and emendation of the family Alicyclobacillaceae da Costa and Rainey, 2010.
journal, September 2011
- Klenk, Hans-Peter; Lapidus, Alla; Chertkov, Olga
- Standards in Genomic Sciences, Vol. 5, Issue 1
Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula
journal, March 2013
- Wegner, Carl-Eric; Richter-Heitmann, Tim; Klindworth, Anna
- Marine Genomics, Vol. 9
The nomenclatural types of the orders Acholeplasmatales, Halanaerobiales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales, Planctomycetales, Prochlorales, Sulfolobales, Thermococcales, Thermoproteales and Verrucomicrobiales are the genera Acholeplasma, Halanaerobium, Halobacterium, Methanobacterium, Methanococcus, Methanomicrobium, Planctomyces, Prochloron, Sulfolobus, Thermococcus, Thermoproteus and Verrucomicrobium, respectively. Opinion 79
journal, January 2005
- Judicial Commission of the International Committee on Systematics of Prokaryotes,
- International Journal of Systematic and Evolutionary Microbiology, Vol. 55, Issue 1, p. 517-518
Validation of the Publication of New Names and New Combinations Previously Effectively Published Outside the IJSB: List No. 23
journal, April 1987
- ,
- International Journal of Systematic Bacteriology, Vol. 37, Issue 2, p. 179-179
Validation of the Publication of New Names and New Combinations Previously Effectively Published Outside the IJSB: List No. 32
journal, January 1990
- ,
- International Journal of Systematic Bacteriology, Vol. 40, Issue 1, p. 105-106
Telmatocola sphagniphila gen. nov., sp. nov., a Novel Dendriform Planctomycete from Northern Wetlands
journal, January 2012
- Kulichevskaya, Irina S.; Serkebaeva, Yulia M.; Kim, Yongkyu
- Frontiers in Microbiology, Vol. 3
The Genome Sequence of Methanohalophilus mahii SLP T Reveals Differences in the Energy Metabolism among Members of the Methanosarcinaceae Inhabiting Freshwater and Saline Environments
journal, January 2010
- Spring, Stefan; Scheuner, Carmen; Lapidus, Alla
- Archaea, Vol. 2010
How Many Bootstrap Replicates Are Necessary?
book, January 2009
- Pattengale, Nicholas D.; Alipour, Masoud; Bininda-Emonds, Olaf R. P.
- Lecture Notes in Computer Science
Gemmata obscuriglobus, a new genus and species of the budding bacteria
journal, May 1984
- Franzmann, P. D.; Skerman, V. B. D.
- Antonie van Leeuwenhoek, Vol. 50, Issue 3
Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell
journal, June 2001
- Lindsay, Margaret; Webb, Richard; Strous, Marc
- Archives of Microbiology, Vol. 175, Issue 6
From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules
journal, August 2013
- Jeske, Olga; Jogler, Mareike; Petersen, Jörn
- Antonie van Leeuwenhoek, Vol. 104, Issue 4
Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula
journal, March 2013
- Wegner, Carl-Eric; Richter-Heitmann, Tim; Klindworth, Anna
- Marine Genomics, Vol. 9
Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses
journal, October 2010
- Yarza, Pablo; Ludwig, Wolfgang; Euzéby, Jean
- Systematic and Applied Microbiology, Vol. 33, Issue 6
The minimum information about a genome sequence (MIGS) specification
journal, May 2008
- Field, Dawn; Garrity, George; Gray, Tanya
- Nature Biotechnology, Vol. 26, Issue 5
Genetic basis for the establishment of endosymbiosis in Paramecium
journal, January 2019
- He, Ming; Wang, Jinfeng; Fan, Xinpeng
- The ISME Journal, Vol. 13, Issue 5
Dietary palmitic acid promotes a prometastatic memory via Schwann cells
journal, November 2021
- Pascual, Gloria; Domínguez, Diana; Elosúa-Bayes, Marc
- Nature, Vol. 599, Issue 7885
Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya.
journal, June 1990
- Woese, C. R.; Kandler, O.; Wheelis, M. L.
- Proceedings of the National Academy of Sciences, Vol. 87, Issue 12
A Rapid Bootstrap Algorithm for the RAxML Web Servers
journal, October 2008
- Stamatakis, Alexandros; Hoover, Paul; Rougemont, Jacques
- Systematic Biology, Vol. 57, Issue 5
The DNA Bank Network: The Start from a German Initiative
journal, March 2011
- Gemeinholzer, Birgit; Dröge, Gabriele; Zetzsche, Holger
- Biopreservation and Biobanking, Vol. 9, Issue 1
RASCAL: rapid scanning and correction of multiple sequence alignments
journal, June 2003
- Thompson, J. D.; Thierry, J. C.; Poch, O.
- Bioinformatics, Vol. 19, Issue 9
opm: an R package for analysing OmniLog(R) phenotype microarray data
journal, June 2013
- Vaas, L. A. I.; Sikorski, J.; Hofner, B.
- Bioinformatics, Vol. 29, Issue 14
A Phylogenomic Approach to Resolve the Arthropod Tree of Life
journal, June 2010
- Meusemann, K.; von Reumont, B. M.; Simon, S.
- Molecular Biology and Evolution, Vol. 27, Issue 11
An efficient algorithm for large-scale detection of protein families
journal, April 2002
- Enright, A. J.
- Nucleic Acids Research, Vol. 30, Issue 7
MUSCLE: multiple sequence alignment with high accuracy and high throughput
journal, March 2004
- Edgar, R. C.
- Nucleic Acids Research, Vol. 32, Issue 5, p. 1792-1797
The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata
journal, December 2011
- Pagani, I.; Liolios, K.; Jansson, J.
- Nucleic Acids Research, Vol. 40, Issue D1
Approved Lists of Bacterial Names
journal, January 1980
- Sneath, P. H. A.; McGOWAN, Vicki; Skerman, V. B. D.
- International Journal of Systematic and Evolutionary Microbiology, Vol. 30, Issue 1
Planctomyces maris sp. nov., nom. rev.
journal, October 1980
- Bauld, J.; Staley, J. T.
- International Journal of Systematic Bacteriology, Vol. 30, Issue 4
Planctomyces stranskae (ex Wawrik 1952) sp. nov., nom. rev. and Planctomyces guttaeformis (ex Hortobagyi 1965) sp. nov., nom. rev.
journal, October 1984
- Starr, M. P.; Schmidt, J. M.
- International Journal of Systematic Bacteriology, Vol. 34, Issue 4
Assignment of ATCC 27377 to Pirella gen. nov. as Pirella staleyi comb. nov.
journal, October 1984
- Schlesner, H.; Hirsch, P.
- International Journal of Systematic Bacteriology, Vol. 34, Issue 4
Phylogenetic diversity, polyamine pattern and DNA base composition of members of the order Planctomycetales
journal, April 1999
- Griepenburg, U.; Ward-Rainey, N.; Mohamed, S.
- International Journal of Systematic Bacteriology, Vol. 49, Issue 2
Planctomyces maris sp. nov.: a Marine Isolate of the Planctomyces-Blastocaulis Group of Budding Bacteria
journal, November 1976
- Bauld, J.; Staley, J. T.
- Journal of General Microbiology, Vol. 97, Issue 1
Singulisphaera rosea sp. nov., a planctomycete from acidic Sphagnum peat, and emended description of the genus Singulisphaera
journal, February 2011
- Kulichevskaya, I. S.; Detkova, E. N.; Bodelier, P. L. E.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 62, Issue 1
Comparative analysis of ribonuclease P RNA of the planctomycetes
journal, July 2004
- Butler, M. K.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 54, Issue 4
Blastopirellula cremea sp. nov., isolated from a dead ark clam
journal, November 2012
- Lee, H. -W.; Roh, S. W.; Shin, N. -R.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 63, Issue Pt 6
Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula
journal, September 2004
- Schlesner, H.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 54, Issue 5
Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from Sphagnum-dominated boreal wetlands
journal, November 2007
- Kulichevskaya, I. S.; Ivanova, A. O.; Belova, S. E.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 57, Issue 11
Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands
journal, May 2008
- Kulichevskaya, I. S.; Ivanova, A. O.; Baulina, O. I.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 58, Issue 5
Parsimony, likelihood, and simplicity
journal, April 2003
- Goloboff, Pablo A.
- Cladistics, Vol. 19, Issue 2
Planktonic microbial community composition across steep physical/chemical gradients in permanently ice-covered Lake Bonney, Antarctica
journal, March 2006
- Glatz, R. E.; Lepp, P. W.; Ward, B. B.
- Geobiology, Vol. 4, Issue 1
Transposon Mutagenesis of Planctomyces limnophilus and Analysis of a pckA Mutant
journal, July 2012
- Schreier, Harold J.; Dejtisakdi, Wipawee; Escalante, Jose Ortega
- Applied and Environmental Microbiology, Vol. 78, Issue 19
Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon
journal, January 1997
- Fuerst, J. A.; Gwilliam, H. G.; Lindsay, M.
- Applied and Environmental Microbiology, Vol. 63, Issue 1
Phylogenetic Systematics
journal, January 1965
- Hennig, Willi
- Annual Review of Entomology, Vol. 10, Issue 1
The Genome Sequence of Methanohalophilus mahii SLP T Reveals Differences in the Energy Metabolism among Members of the Methanosarcinaceae Inhabiting Freshwater and Saline Environments
journal, January 2010
- Spring, Stefan; Scheuner, Carmen; Lapidus, Alla
- Archaea, Vol. 2010
Novel Insights into the Diversity of Catabolic Metabolism from Ten Haloarchaeal Genomes
journal, May 2011
- Anderson, Iain; Scheuner, Carmen; Göker, Markus
- PLoS ONE, Vol. 6, Issue 5
Codivergence of Mycoviruses with Their Hosts
journal, July 2011
- Göker, Markus; Scheuner, Carmen; Klenk, Hans-Peter
- PLoS ONE, Vol. 6, Issue 7
The Fast Changing Landscape of Sequencing Technologies and Their Impact on Microbial Genome Assemblies and Annotation
journal, December 2012
- Mavromatis, Konstantinos; Land, Miriam L.; Brettin, Thomas S.
- PLoS ONE, Vol. 7, Issue 12
Genomic Evolution of 11 Type Strains within Family Planctomycetaceae
journal, January 2014
- Guo, Min; Zhou, Qian; Zhou, Yizhuang
- PLoS ONE, Vol. 9, Issue 1
The Information Content of the Phylogenetic System
journal, December 1979
- Farris, J. S.
- Systematic Biology, Vol. 28, Issue 4
Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes
journal, January 2009
- Fukunaga, Yukiyo; Kurahashi, Midori; Sakiyama, Yayoi
- The Journal of General and Applied Microbiology, Vol. 55, Issue 4
Complete genome sequence of Planctomyces limnophilus type strain (Mü 290T)
journal, July 2010
- LaButti, Kurt; Sikorski, Johannes; Schneider, Susanne
- Standards in Genomic Sciences, Vol. 3, Issue 1
Complete genome sequence of Isosphaera pallida type strain (IS1BT)
journal, February 2011
- Göker, Markus; Cleland, David; Saunders, Elizabeth
- Standards in Genomic Sciences, Vol. 4, Issue 1
Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21T)
journal, October 2011
- Chang, Yun-juan; Land, Miriam; Hauser, Loren
- Standards in Genomic Sciences, Vol. 5, Issue 1
Complete genome sequence of the termite hindgut bacterium Spirochaeta coccoides type strain (SPN1T), reclassification in the genus Sphaerochaeta as Sphaerochaeta coccoides comb. nov. and emendations of the family Spirochaetaceae and the genus Sphaerochaeta
journal, May 2012
- Abt, Birte; Han, Cliff; Scheuner, Carmen
- Standards in Genomic Sciences, Vol. 6, Issue 2
Phylogeny-driven target selection for large-scale genome-sequencing (and other) projects
journal, May 2013
- Göker, Markus; Klenk, Hans-Peter
- Standards in Genomic Sciences, Vol. 8, Issue 2
Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison
journal, January 2010
- Auch, Alexander F.; von Jan, Mathias; Klenk, Hans-Peter
- Standards in Genomic Sciences, Vol. 2, Issue 1
Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs
journal, January 2010
- Auch, Alexander F.; Klenk, Hans-Peter; Göker, Markus
- Standards in Genomic Sciences, Vol. 2, Issue 1
Complete genome sequence of Kytococcus sedentarius type strain (541T)
journal, July 2009
- Sims, David; Brettin, Thomas; Detter, John C.
- Standards in Genomic Sciences, Vol. 1, Issue 1
Works referencing / citing this record:
Global and Targeted Lipid Analysis of Gemmata obscuriglobus Reveals the Presence of Lipopolysaccharide, a Signature of the Classical Gram-Negative Outer Membrane
journal, October 2015
- Mahat, Rajendra; Seebart, Corrine; Basile, Franco
- Journal of Bacteriology, Vol. 198, Issue 2
Biochar-induced changes in metal mobility and uptake by perennial plants in a ferralsol of Brazil’s Atlantic forest
journal, July 2019
- von Gunten, Konstantin; Hubmann, Magdalena; Ineichen, Robert
- Biochar, Vol. 1, Issue 3
Comparative genomics of biotechnologically important yeasts
journal, August 2016
- Riley, Robert; Haridas, Sajeet; Wolfe, Kenneth H.
- Proceedings of the National Academy of Sciences, Vol. 113, Issue 35
On the maverick Planctomycetes
journal, July 2018
- Wiegand, Sandra; Jogler, Mareike; Jogler, Christian
- FEMS Microbiology Reviews, Vol. 42, Issue 6
Limnoglobus roseus gen. nov., sp. nov., a novel freshwater planctomycete with a giant genome from the family Gemmataceae
journal, February 2020
- Kulichevskaya, Irina S.; Naumoff, Daniil G.; Miroshnikov, Kirill K.
- International Journal of Systematic and Evolutionary Microbiology, Vol. 70, Issue 2
Genome-Based Taxonomic Classification of Bacteroidetes
journal, December 2016
- Hahnke, Richard L.; Meier-Kolthoff, Jan P.; García-López, Marina
- Frontiers in Microbiology, Vol. 7
Rubinisphaera italica sp. nov. isolated from a hydrothermal area in the Tyrrhenian Sea close to the volcanic island Panarea
journal, November 2019
- Kallscheuer, Nicolai; Jogler, Mareike; Wiegand, Sandra
- Antonie van Leeuwenhoek
Fuerstia marisgermanicae gen. nov., sp. nov., an Unusual Member of the Phylum Planctomycetes from the German Wadden Sea
journal, December 2016
- Kohn, Timo; Heuer, Anja; Jogler, Mareike
- Frontiers in Microbiology, Vol. 7
Pink‐ and orange‐pigmented Planctomycetes produce saproxanthin‐type carotenoids including a rare C 45 carotenoid
journal, October 2019
- Kallscheuer, Nicolai; Moreira, Catia; Airs, Ruth
- Environmental Microbiology Reports
Comparative Genomics of Four Isosphaeraceae Planctomycetes: A Common Pool of Plasmids and Glycoside Hydrolase Genes Shared by Paludisphaera borealis PX4T, Isosphaera pallida IS1BT, Singulisphaera acidiphila DSM 18658T, and Strain SH-PL62
journal, March 2017
- Ivanova, Anastasia A.; Naumoff, Daniil G.; Miroshnikov, Kirill K.
- Frontiers in Microbiology, Vol. 8
Alienimonas californiensis gen. nov. sp. nov., a novel Planctomycete isolated from the kelp forest in Monterey Bay
journal, December 2019
- Boersma, Alje S.; Kallscheuer, Nicolai; Wiegand, Sandra
- Antonie van Leeuwenhoek
100‐year‐old enigma solved: identification, genomic characterization and biogeography of the yet uncultured Planctomyces bekefii
journal, November 2019
- Dedysh, Svetlana N.; Henke, Petra; Ivanova, Anastasia A.
- Environmental Microbiology, Vol. 22, Issue 1
Tuwongella immobilis gen. nov., sp. nov., a novel non-motile bacterium within the phylum Planctomycetes
journal, December 2017
- Seeger, Christian; Butler, Margaret K.; Yee, Benjamin
- International Journal of Systematic and Evolutionary Microbiology, Vol. 67, Issue 12
Genome-based classification of micromonosporae with a focus on their biotechnological and ecological potential
journal, January 2018
- Carro, Lorena; Nouioui, Imen; Sangal, Vartul
- Scientific Reports, Vol. 8, Issue 1
Draft genome sequences of Bradyrhizobium shewense sp. nov. ERR11T and Bradyrhizobium yuanmingense CCBAU 10071T
journal, December 2017
- Aserse, Aregu Amsalu; Woyke, Tanja; Kyrpides, Nikos C.
- Standards in Genomic Sciences, Vol. 12, Issue 1
Assembly of a complete genome sequence for Gemmata obscuriglobus reveals a novel prokaryotic rRNA operon gene architecture
journal, May 2018
- Franke, Josef D.; Blomberg, Wilson R.; Todd, Robert T.
- Antonie van Leeuwenhoek, Vol. 111, Issue 11
Exemplar Abstract for Planctomyces brasiliensis Schlesner 1990 and Rubinisphaera brasiliensis (Schlesner 1990) Scheuner et al. 2015.
dataset, January 2013
- Parker, Charles Thomas; Taylor, Dorothea; Garrity, George M.
Exemplar Abstract for Planctomyces limnophilus Hirsch and M??ller 1986, Planctopirus limnophila corrig. (Hirsch and M??ller 1986) Scheuner et al. 2015 and Planctopirus limnophilus (sic) (Hirsch and M??ller 1986) Scheuner et al. 2015.
dataset, January 2013
- Parker, Charles Thomas; Taylor, Dorothea; Garrity, George M.
Nomenclature Abstract for Singulisphaera acidiphila Kulichevskaya et al. 2008 emend. Scheuner et al. 2014.
dataset, January 2013
- Parker, Charles Thomas; Wigley, Sarah; Garrity, George M.
Nomenclature Abstract for Rubinisphaera Scheuner et al. 2015 emend. Kallscheuer et al. 2020.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Nomenclature Abstract for Rubinisphaera brasiliensis (Schlesner 1990) Scheuner et al. 2015.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Nomenclature Abstract for Planctopirus Scheuner et al. 2015.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Nomenclature Abstract for Planctopirus limnophilus (sic) (Hirsch and Muller 1986) Scheuner et al. 2015.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Nomenclature Abstract for Gimesia Scheuner et al. 2015 emend. Kumar et al. 2020.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Nomenclature Abstract for Gimesia maris (Bauld and Staley 1980) Scheuner et al. 2015.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Nomenclature Abstract for Planctopirus limnophila corrig. (Hirsch and Muller 1986) Scheuner et al. 2015.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Nomenclature Abstract for Planctomycetales Schlesner and Stackebrandt 1987 emend. Dedysh et al. 2020.
dataset, January 2013
- Parker, Charles Thomas; Wigley, Sarah; Garrity, George M.
Nomenclature Abstract for Planctomycetaceae Schlesner and Stackebrandt 1987 emend. Dedysh et al. 2020.
dataset, January 2013
- Parker, Charles Thomas; Wigley, Sarah; Garrity, George M.
Nomenclature Abstract for Planctomyces brasiliensis Schlesner 1990.
dataset, January 2013
- Parker, Charles Thomas; Osier, Nicole Danielle; Garrity, George M.
Nomenclature Abstract for Planctomyces limnophilus Hirsch and Muller 1986.
dataset, January 2013
- Parker, Charles Thomas; Osier, Nicole Danielle; Garrity, George M.
Nomenclature Abstract for Planctomyces maris (ex Bauld and Staley 1976) Bauld and Staley 1980.
dataset, January 2013
- Parker, Charles Thomas; Osier, Nicole Danielle; Garrity, George M.
Nomenclature Abstract for Gemmata Franzmann and Skerman 1985 emend. Scheuner et al. 2014.
dataset, January 2013
- Parker, Charles Thomas; Garrity, George M.
Taxonomic Abstract for the species.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Taxonomic Abstract for the species.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Taxonomic Abstract for the species.
dataset, January 2015
- Parker, Charles Thomas; Garrity, George M.
Taxonomic Abstract for the families.
dataset, January 2013
- Parker, Charles Thomas; Wigley, Sarah; Garrity, George M.
Genome-based classification of micromonosporae with a focus on their biotechnological and ecological potential
journal, January 2018
- Carro, Lorena; Nouioui, Imen; Sangal, Vartul
- Scientific Reports, Vol. 8, Issue 1
A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data
journal, April 2015
- Spring, Stefan; Scheuner, Carmen; Göker, Markus
- Frontiers in Microbiology, Vol. 6
Genome-Based Taxonomic Classification of Bacteroidetes
journal, December 2016
- Hahnke, Richard L.; Meier-Kolthoff, Jan P.; García-López, Marina
- Frontiers in Microbiology, Vol. 7
Fuerstia marisgermanicae gen. nov., sp. nov., an Unusual Member of the Phylum Planctomycetes from the German Wadden Sea
journal, December 2016
- Kohn, Timo; Heuer, Anja; Jogler, Mareike
- Frontiers in Microbiology, Vol. 7
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal