Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants
Abstract
Background: Prasinophytes are widespread marine green algae that are related to plants. Cellular abundance of the prasinophyte Micromonas has reportedly increased in the Arctic due to climate-induced changes. Thus, studies of these unicellular eukaryotes are important for marine ecology and for understanding Viridiplantae evolution and diversification. Results: We generated evidence-based Micromonas gene models using proteomics and RNA-Seq to improve prasinophyte genomic resources. First, sequences of four chromosomes in the 22 Mb Micromonas pusilla (CCMP1545) genome were finished. Comparison with the finished 21 Mb genome of Micromonas commoda (RCC299; named herein) shows they share ≤8,141 of ~10,000 protein-encoding genes, depending on the analysis method. Unlike RCC299 and other sequenced eukaryotes, CCMP1545 has two abundant repetitive intron types and a high percent (26 %) GC splice donors. Micromonas has more genus-specific protein families (19 %) than other genome sequenced prasinophytes (11 %). Comparative analyses using predicted proteomes from other prasinophytes reveal proteins likely related to scale formation and ancestral photosynthesis. Our studies also indicate that peptidoglycan (PG) biosynthesis enzymes have been lost in multiple independent events in select prasinophytes and plants. However, CCMP1545, polar Micromonas CCMP2099 and prasinophytes from other classes retain the entire PG pathway, like moss and glaucophyte algae. Surprisingly,more »
- Authors:
- Publication Date:
- Research Org.:
- Pacific Northwest National Laboratory (PNNL), Richland, WA (United States). Environmental Molecular Sciences Laboratory (EMSL); Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC), Biological and Environmental Research (BER)
- OSTI Identifier:
- 1618541
- Alternate Identifier(s):
- OSTI ID: 1337274; OSTI ID: 1615262
- Report Number(s):
- PNNL-SA-114561
Journal ID: ISSN 1471-2164; 267; PII: 2585
- Grant/Contract Number:
- SC0004765; AC05-76RL01830; AC02-05CH11231
- Resource Type:
- Published Article
- Journal Name:
- BMC Genomics
- Additional Journal Information:
- Journal Name: BMC Genomics Journal Volume: 17 Journal Issue: 1; Journal ID: ISSN 1471-2164
- Publisher:
- Springer
- Country of Publication:
- United Kingdom
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES; Environmental Molecular Sciences Laboratory; GreenCut; Archaeplastida evolution; Viridiplantae; Introner Elements; RNA sequencing; Proteomics; Evidence-based gene models; Peptidoglycan; PPASP
Citation Formats
van Baren, Marijke J., Bachy, Charles, Reistetter, Emily Nahas, Purvine, Samuel O., Grimwood, Jane, Sudek, Sebastian, Yu, Hang, Poirier, Camille, Deerinck, Thomas J., Kuo, Alan, Grigoriev, Igor V., Wong, Chee-Hong, Smith, Richard D., Callister, Stephen J., Wei, Chia-Lin, Schmutz, Jeremy, and Worden, Alexandra Z. Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants. United Kingdom: N. p., 2016.
Web. doi:10.1186/s12864-016-2585-6.
van Baren, Marijke J., Bachy, Charles, Reistetter, Emily Nahas, Purvine, Samuel O., Grimwood, Jane, Sudek, Sebastian, Yu, Hang, Poirier, Camille, Deerinck, Thomas J., Kuo, Alan, Grigoriev, Igor V., Wong, Chee-Hong, Smith, Richard D., Callister, Stephen J., Wei, Chia-Lin, Schmutz, Jeremy, & Worden, Alexandra Z. Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants. United Kingdom. https://doi.org/10.1186/s12864-016-2585-6
van Baren, Marijke J., Bachy, Charles, Reistetter, Emily Nahas, Purvine, Samuel O., Grimwood, Jane, Sudek, Sebastian, Yu, Hang, Poirier, Camille, Deerinck, Thomas J., Kuo, Alan, Grigoriev, Igor V., Wong, Chee-Hong, Smith, Richard D., Callister, Stephen J., Wei, Chia-Lin, Schmutz, Jeremy, and Worden, Alexandra Z. Thu .
"Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants". United Kingdom. https://doi.org/10.1186/s12864-016-2585-6.
@article{osti_1618541,
title = {Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants},
author = {van Baren, Marijke J. and Bachy, Charles and Reistetter, Emily Nahas and Purvine, Samuel O. and Grimwood, Jane and Sudek, Sebastian and Yu, Hang and Poirier, Camille and Deerinck, Thomas J. and Kuo, Alan and Grigoriev, Igor V. and Wong, Chee-Hong and Smith, Richard D. and Callister, Stephen J. and Wei, Chia-Lin and Schmutz, Jeremy and Worden, Alexandra Z.},
abstractNote = {Background: Prasinophytes are widespread marine green algae that are related to plants. Cellular abundance of the prasinophyte Micromonas has reportedly increased in the Arctic due to climate-induced changes. Thus, studies of these unicellular eukaryotes are important for marine ecology and for understanding Viridiplantae evolution and diversification. Results: We generated evidence-based Micromonas gene models using proteomics and RNA-Seq to improve prasinophyte genomic resources. First, sequences of four chromosomes in the 22 Mb Micromonas pusilla (CCMP1545) genome were finished. Comparison with the finished 21 Mb genome of Micromonas commoda (RCC299; named herein) shows they share ≤8,141 of ~10,000 protein-encoding genes, depending on the analysis method. Unlike RCC299 and other sequenced eukaryotes, CCMP1545 has two abundant repetitive intron types and a high percent (26 %) GC splice donors. Micromonas has more genus-specific protein families (19 %) than other genome sequenced prasinophytes (11 %). Comparative analyses using predicted proteomes from other prasinophytes reveal proteins likely related to scale formation and ancestral photosynthesis. Our studies also indicate that peptidoglycan (PG) biosynthesis enzymes have been lost in multiple independent events in select prasinophytes and plants. However, CCMP1545, polar Micromonas CCMP2099 and prasinophytes from other classes retain the entire PG pathway, like moss and glaucophyte algae. Surprisingly, multiple vascular plants also have the PG pathway, except the Penicillin-Binding Protein, and share a unique bi-domain protein potentially associated with the pathway. Alongside Micromonas experiments using antibiotics that halt bacterial PG biosynthesis, the findings highlight unrecognized phylogenetic complexity in PG-pathway retention and implicate a role in chloroplast structure or division in several extant Viridiplantae lineages. Conclusions: Extensive differences in gene loss and architecture between related prasinophytes underscore their divergence. PG biosynthesis genes from the cyanobacterial endosymbiont that became the plastid, have been selectively retained in multiple plants and algae, implying a biological function. Our studies provide robust genomic resources for emerging model algae, advancing knowledge of marine phytoplankton and plant evolution},
doi = {10.1186/s12864-016-2585-6},
journal = {BMC Genomics},
number = 1,
volume = 17,
place = {United Kingdom},
year = {Thu Mar 31 00:00:00 EDT 2016},
month = {Thu Mar 31 00:00:00 EDT 2016}
}
https://doi.org/10.1186/s12864-016-2585-6
Web of Science
Figures / Tables:
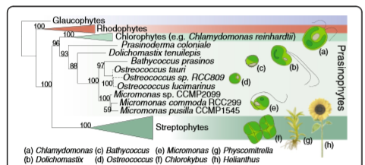 Fig. 1: Relationships of photosynthetic eukaryotes in the Archaeplastida. The cladogram was inferred with the cpREV+G model using maximum likelihood methods and a concatenated alignment of 16 plastid-genome encoded proteins. Bootstrap support >50 % is shown. Plastid genome information is lacking for O. lucimarinus and RCC809, therefore, branching within themore »
Fig. 1: Relationships of photosynthetic eukaryotes in the Archaeplastida. The cladogram was inferred with the cpREV+G model using maximum likelihood methods and a concatenated alignment of 16 plastid-genome encoded proteins. Bootstrap support >50 % is shown. Plastid genome information is lacking for O. lucimarinus and RCC809, therefore, branching within themore »
Works referenced in this record:
The Mechanism of Action of Fosfomycin (Phosphonomycin)
journal, May 1974
- Kahan, Frederick M.; Kahan, Jean S.; Cassidy, Patrick J.
- Annals of the New York Academy of Sciences, Vol. 235, Issue 1 Mode of Actio
FIMO: scanning for occurrences of a given motif
journal, February 2011
- Grant, Charles E.; Bailey, Timothy L.; Noble, William Stafford
- Bioinformatics, Vol. 27, Issue 7
The voyage of the microbial eukaryote
journal, October 2010
- Worden, Alexandra Z.; Allen, Andrew E.
- Current Opinion in Microbiology, Vol. 13, Issue 5
New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0
journal, March 2010
- Guindon, Stéphane; Dufayard, Jean-François; Lefort, Vincent
- Systematic Biology, Vol. 59, Issue 3
MEME: discovering and analyzing DNA and protein sequence motifs
journal, July 2006
- Bailey, T. L.; Williams, N.; Misleh, C.
- Nucleic Acids Research, Vol. 34, Issue Web Server
Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis
journal, April 2000
- Castresana, J.
- Molecular Biology and Evolution, Vol. 17, Issue 4
Contrasting Strategies of Photosynthetic Energy Utilization Drive Lifestyle Strategies in Ecologically Important Picoeukaryotes
journal, April 2014
- Halsey, Kimberly; Milligan, Allen; Behrenfeld, Michael
- Metabolites, Vol. 4, Issue 2
Alternatives to vitamin B1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages
journal, August 2014
- McRose, Darcy; Guo, Jian; Monier, Adam
- The ISME Journal, Vol. 8, Issue 12
The ultrastructure of Bathycoccus gen. nov. and B. prasinos sp. nov., a non-motile picoplanktonic alga (Chlorophyta, Prasinophyceae) from the Mediterranean and Atlantic
journal, September 1990
- Eikrem, W.; Throndsen, J.
- Phycologia, Vol. 29, Issue 3
Into the deep: New discoveries at the base of the green plant phylogeny
journal, July 2011
- Leliaert, Frederik; Verbruggen, Heroen; Zechman, Frederick W.
- BioEssays, Vol. 33, Issue 9
Evidence for Extensive Recent Intron Transposition in Closely Related Fungi
journal, December 2011
- Torriani, Stefano F. F.; Stukenbrock, Eva H.; Brunner, Patrick C.
- Current Biology, Vol. 21, Issue 23
InterProScan - an integration platform for the signature-recognition methods in InterPro
journal, September 2001
- Zdobnov, E. M.; Apweiler, R.
- Bioinformatics, Vol. 17, Issue 9, p. 847-848
Treatment with Antibiotics that Interfere with Peptidoglycan Biosynthesis Inhibits Chloroplast Division in the Desmid Closterium
journal, July 2012
- Matsumoto, Hiroko; Takechi, Katsuaki; Sato, Hiroshi
- PLoS ONE, Vol. 7, Issue 7
Picoeukaryote diversity in coastal waters of the Pacific Ocean
journal, June 2006
- Worden, Az
- Aquatic Microbial Ecology, Vol. 43
Birth of New Spliceosomal Introns in Fungi by Multiplication of Introner-like Elements
journal, July 2012
- van der Burgt, Ate; Severing, Edouard; de Wit, Pierre J. G. M.
- Current Biology, Vol. 22, Issue 13
Chemically Etched Open Tubular and Monolithic Emitters for Nanoelectrospray Ionization Mass Spectrometry
journal, November 2006
- Kelly, Ryan T.; Page, Jason S.; Luo, Quanzhou
- Analytical Chemistry, Vol. 78, Issue 22, p. 7796-7801
FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments
journal, March 2010
- Price, Morgan N.; Dehal, Paramvir S.; Arkin, Adam P.
- PLoS ONE, Vol. 5, Issue 3
Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks
journal, March 2012
- Trapnell, Cole; Roberts, Adam; Goff, Loyal
- Nature Protocols, Vol. 7, Issue 3
The Complex Intron Landscape and Massive Intron Invasion in a Picoeukaryote Provides Insights into Intron Evolution
journal, November 2013
- Verhelst, Bram; Van de Peer, Yves; Rouzé, Pierre
- Genome Biology and Evolution, Vol. 5, Issue 12
Non-random retention of protein-coding overlapping genes in Metazoa
journal, January 2008
- Soldà, Giulia; Suyama, Mikita; Pelucchi, Paride
- BMC Genomics, Vol. 9, Issue 1
Analysis of Biostimulated Microbial Communities from Two Field Experiments Reveals Temporal and Spatial Differences in Proteome Profiles
journal, December 2010
- Callister, Stephen J.; Wilkins, Michael J.; Nicora, Carrie D.
- Environmental Science & Technology, Vol. 44, Issue 23
Sel1-like repeat proteins in signal transduction
journal, January 2007
- Mittl, Peer R. E.; Schneider-Brachert, Wulf
- Cellular Signalling, Vol. 19, Issue 1
Full-length transcriptome assembly from RNA-Seq data without a reference genome
journal, May 2011
- Grabherr, Manfred G.; Haas, Brian J.; Yassour, Moran
- Nature Biotechnology, Vol. 29, Issue 7
Ecological niche partitioning in the picoplanktonic green alga Micromonas pusilla : evidence from environmental surveys using phylogenetic probes
journal, September 2008
- Foulon, Elodie; Not, Fabrice; Jalabert, Fabienne
- Environmental Microbiology, Vol. 10, Issue 9
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
journal, January 2013
- Katoh, K.; Standley, D. M.
- Molecular Biology and Evolution, Vol. 30, Issue 4
Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage
journal, January 2012
- Moreau, Hervé; Verhelst, Bram; Couloux, Arnaud
- Genome Biology, Vol. 13, Issue 8
The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) 1 1Edited by P. E. Wight
journal, June 2000
- Bateman, Alex; Bycroft, Mark
- Journal of Molecular Biology, Vol. 299, Issue 4
The Peptidoglycan Biosynthesis Genes MurA and MraY are Related to Chloroplast Division in the Moss Physcomitrella patens
journal, September 2009
- Homi, Shoko; Takechi, Katsuaki; Tanidokoro, Koji
- Plant and Cell Physiology, Vol. 50, Issue 12
Dolichomastix tenuilepis sp. nov., a first insight into the microanatomy of the genus Dolichomastix (Mamiellales, Prasinophyceae, Chlorophyta)
journal, May 1997
- Throndsen, Jahn; Zingone, Adriana
- Phycologia, Vol. 36, Issue 3
The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions
journal, October 2007
- Merchant, S. S.; Prochnik, S. E.; Vallon, O.
- Science, Vol. 318, Issue 5848, p. 245-250
Where will new antibiotics come from?
journal, October 2003
- Walsh, Christopher
- Nature Reviews Microbiology, Vol. 1, Issue 1
The UCSC Genome Browser database: extensions and updates 2011
journal, November 2011
- Dreszer, T. R.; Karolchik, D.; Zweig, A. S.
- Nucleic Acids Research, Vol. 40, Issue D1
Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss
journal, April 2006
- Machida, M.; Takechi, K.; Sato, H.
- Proceedings of the National Academy of Sciences, Vol. 103, Issue 17
Xyloglucan Fucosyltransferase, an Enzyme Involved in Plant Cell Wall Biosynthesis
journal, June 1999
- Perrin, R. M.
- Science, Vol. 284, Issue 5422
Contributions to our knowledge of the smaller marine algae
journal, June 1952
- Butcher, R. W.
- Journal of the Marine Biological Association of the United Kingdom, Vol. 31, Issue 1
The GreenCut2 Resource, a Phylogenomically Derived Inventory of Proteins Specific to the Plant Lineage
journal, April 2011
- Karpowicz, Steven J.; Prochnik, Simon E.; Grossman, Arthur R.
- Journal of Biological Chemistry, Vol. 286, Issue 24
Structural analysis of the flagellar apparatus and the scaly periplast in Chrysochromulina scutellum sp. nov. (Prymnesiophyceae, Haptophyta) from the Skagerrak and the Baltic
journal, March 1998
- Eikrem, Wenche; Moestrup, Øjvind
- Phycologia, Vol. 37, Issue 2
Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components
journal, July 1998
- Field, C. B.
- Science, Vol. 281, Issue 5374
Global Dispersal and Ancient Cryptic Species in the Smallest Marine Eukaryotes
journal, August 2005
- Šlapeta, Jan; López-García, Purificación; Moreira, David
- Molecular Biology and Evolution, Vol. 23, Issue 1
The diversity of small eukaryotic phytoplankton (≤3 μm) in marine ecosystems
journal, August 2008
- Vaulot, Daniel; Eikrem, Wenche; Viprey, Manon
- FEMS Microbiology Reviews, Vol. 32, Issue 5
Dolichomastix (Prasinophyceae) from arctic Canada, Alaska and South Africa: a new genus of flagellates with scaly flagella
journal, December 1977
- Manton, I.
- Phycologia, Vol. 16, Issue 4
Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen
journal, January 2001
- Krogh, Anders; Larsson, Björn; von Heijne, Gunnar
- Journal of Molecular Biology, Vol. 305, Issue 3
Characterization of individual mouse cerebrospinal fluid proteomes
journal, March 2014
- Smith, Jeffrey S.; Angel, Thomas E.; Chavkin, Charles
- PROTEOMICS, Vol. 14, Issue 9
Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence
journal, July 2000
- Emanuelsson, Olof; Nielsen, Henrik; Brunak, Søren
- Journal of Molecular Biology, Vol. 300, Issue 4
Plastid Genome Phylogeny and a Model of Amino Acid Substitution for Proteins Encoded by Chloroplast DNA
journal, April 2000
- Adachi, Jun; Waddell, Peter J.; Martin, William
- Journal of Molecular Evolution, Vol. 50, Issue 4
Consed: a graphical editor for next-generation sequencing
journal, August 2013
- Gordon, David; Green, Phil
- Bioinformatics, Vol. 29, Issue 22
Pan genome of the phytoplankton Emiliania underpins its global distribution
journal, June 2013
- Read, Betsy A.; Kegel, Jessica; Klute, Mary J.
- Nature, Vol. 499, Issue 7457
Plasticity of Animal Genome Architecture Unmasked by Rapid Evolution of a Pelagic Tunicate
journal, November 2010
- Denoeud, F.; Henriet, S.; Mungpakdee, S.
- Science, Vol. 330, Issue 6009
The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation
journal, April 2007
- Palenik, B.; Grimwood, J.; Aerts, A.
- Proceedings of the National Academy of Sciences, Vol. 104, Issue 18
Studies on scale morphogenesis in the Golgi apparatus of Pyramimonas tetrarhynchus (Prasinophyceae)
journal, April 1979
- Moestrup, O.; Walne, P. L.
- Journal of Cell Science, Vol. 36, Issue 1
Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa
journal, January 2013
- Tong, Chaobo; Wang, Xiaowu; Yu, Jingyin
- BMC Genomics, Vol. 14, Issue 1
Self-made frits for nanoscale columns in proteomics
journal, October 2005
- Maiolica, Alessio; Borsotti, Dario; Rappsilber, Juri
- PROTEOMICS, Vol. 5, Issue 15
Xyloglucans in the Primary Cell Wall
journal, June 1989
- Hayashi, T.
- Annual Review of Plant Physiology and Plant Molecular Biology, Vol. 40, Issue 1
Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus
journal, February 2011
- Demir-Hilton, Elif; Sudek, Sebastian; Cuvelier, Marie L.
- The ISME Journal, Vol. 5, Issue 7
Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model
journal, July 2014
- Hengel, Shawna; Aldrich, Joshua; Waters, Katrina
- Proteomes, Vol. 2, Issue 3
Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry
journal, February 2007
- Elias, Joshua E.; Gygi, Steven P.
- Nature Methods, Vol. 4, Issue 3
Marine algae and land plants share conserved phytochrome signaling systems
journal, September 2014
- Duanmu, D.; Bachy, C.; Sudek, S.
- Proceedings of the National Academy of Sciences, Vol. 111, Issue 44
Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection
journal, November 2011
- Willmann, R.; Lajunen, H. M.; Erbs, G.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 49
Phylogenomic Evidence for Separate Acquisition of Plastids in Cryptophytes, Haptophytes, and Stramenopiles
journal, March 2010
- Baurain, D.; Brinkmann, H.; Petersen, J.
- Molecular Biology and Evolution, Vol. 27, Issue 7
MUST, a computer package of Management Utilities for Sequences and Trees
journal, January 1993
- Philippe, Hervé
- Nucleic Acids Research, Vol. 21, Issue 22
TopHat: discovering splice junctions with RNA-Seq
journal, March 2009
- Trapnell, Cole; Pachter, Lior; Salzberg, Steven L.
- Bioinformatics, Vol. 25, Issue 9
DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra
journal, February 2008
- Mayampurath, A. M.; Jaitly, N.; Purvine, S. O.
- Bioinformatics, Vol. 24, Issue 7
Gene invasion in distant eukaryotic lineages: discovery of mutually exclusive genetic elements reveals marine biodiversity
journal, May 2013
- Monier, Adam; Sudek, Sebastian; Fast, Naomi M.
- The ISME Journal, Vol. 7, Issue 9
Characterization of a Family of Arabidopsis Genes Related to Xyloglucan Fucosyltransferase1
journal, December 2001
- Sarria, Rodrigo; Wagner, Tanya A.; O'Neill, Malcolm A.
- Plant Physiology, Vol. 127, Issue 4
Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features
journal, July 2006
- Derelle, E.; Ferraz, C.; Rombauts, S.
- Proceedings of the National Academy of Sciences, Vol. 103, Issue 31
The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis
journal, March 2008
- Sauvage, Eric; Kerff, Frédéric; Terrak, Mohammed
- FEMS Microbiology Reviews, Vol. 32, Issue 2
Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae): Ecotype diversity in the picoeukaryote Ostreococus
journal, March 2005
- Rodríguez, Francisco; Derelle, Evelyne; Guillou, Laure
- Environmental Microbiology, Vol. 7, Issue 6
PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium
journal, December 2009
- Mi, Huaiyu; Dong, Qing; Muruganujan, Anushya
- Nucleic Acids Research, Vol. 38, Issue suppl_1
Phytozome: a comparative platform for green plant genomics
journal, November 2011
- Goodstein, David M.; Shu, Shengqiang; Howson, Russell
- Nucleic Acids Research, Vol. 40, Issue D1
An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database
journal, November 1994
- Eng, Jimmy K.; McCormack, Ashley L.; Yates, John R.
- Journal of the American Society for Mass Spectrometry, Vol. 5, Issue 11
Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags
journal, August 2002
- Lipton, M. S.; Pasa-Tolic, L.; Anderson, G. A.
- Proceedings of the National Academy of Sciences, Vol. 99, Issue 17
LysM, a widely distributed protein motif for binding to (peptido)glycans
journal, May 2008
- Buist, Girbe; Steen, Anton; Kok, Jan
- Molecular Microbiology, Vol. 68, Issue 4
Phosphate transporters in marine phytoplankton and their viruses: cross-domain commonalities in viral-host gene exchanges: Phosphate and commonalities in marine viral-host exchanges
journal, September 2011
- Monier, Adam; Welsh, Rory M.; Gentemann, Chelle
- Environmental Microbiology, Vol. 14, Issue 1
The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing
journal, June 2014
- Keeling, Patrick J.; Burki, Fabien; Wilcox, Heather M.
- PLoS Biology, Vol. 12, Issue 6
OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes
journal, September 2003
- Li, L.
- Genome Research, Vol. 13, Issue 9
Media for the Culture of Oceanic Ultraphytoplankton1,2
journal, December 1987
- Keller, Maureen D.; Selvin, Rhonda C.; Claus, Wolfgang
- Journal of Phycology, Vol. 23, Issue 4
Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic Oceans
journal, February 2014
- McKie-Krisberg, Zaid M.; Sanders, Robert W.
- The ISME Journal, Vol. 8, Issue 10
Characterizing Microbial Community and Geochemical Dynamics at Hydrothermal Vents Using Osmotically Driven Continuous Fluid Samplers
journal, April 2013
- Robidart, Julie; Callister, Stephen J.; Song, Pengfei
- Environmental Science & Technology, Vol. 47, Issue 9
BLAT---The BLAST-Like Alignment Tool
journal, March 2002
- Kent, W. J.
- Genome Research, Vol. 12, Issue 4
The biosynthesis of peptidoglycan lipid-linked intermediates
journal, March 2008
- Bouhss, Ahmed; Trunkfield, Amy E.; Bugg, Timothy D. H.
- FEMS Microbiology Reviews, Vol. 32, Issue 2
Evolution of xyloglucan-related genes in green plants
journal, January 2010
- Del Bem, Luiz; Vincentz, Michel GA
- BMC Evolutionary Biology, Vol. 10, Issue 1
Molecular Phylogeny and Classification of the Mamiellophyceae class. nov. (Chlorophyta) based on Sequence Comparisons of the Nuclear- and Plastid-encoded rRNA Operons
journal, April 2010
- Marin, Birger; Melkonian, Michael
- Protist, Vol. 161, Issue 2
An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development: MurE is essential for chloroplast development
journal, November 2007
- Garcia, Marlon; Myouga, Fumiyoshi; Takechi, Katsuaki
- The Plant Journal, Vol. 53, Issue 6
Green Evolution and Dynamic Adaptations Revealed by Genomes of the Marine Picoeukaryotes Micromonas
journal, April 2009
- Worden, A. Z.; Lee, J. -H.; Mock, T.
- Science, Vol. 324, Issue 5924
Smallest Algae Thrive As the Arctic Ocean Freshens
journal, October 2009
- Li, W. K. W.; McLaughlin, F. A.; Lovejoy, C.
- Science, Vol. 326, Issue 5952
Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component
journal, January 2004
- Worden, Alexandra Z.; Nolan, Jessica K.; Palenik, B.
- Limnology and Oceanography, Vol. 49, Issue 1
Effects of Antibiotics that Inhibit the Bacterial Peptidoglycan Synthesis Pathway on Moss Chloroplast Division
journal, July 2003
- Katayama, Nami; Takano, Hiroyoshi; Sugiyama, Motoji
- Plant and Cell Physiology, Vol. 44, Issue 7
Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles
journal, January 2010
- Monnier, Annabelle; Liverani, Silvia; Bouvet, Regis
- BMC Genomics, Vol. 11, Issue 1
Intron Invasions Trace Algal Speciation and Reveal Nearly Identical Arctic and Antarctic Micromonas Populations
journal, May 2015
- Simmons, Melinda P.; Bachy, Charles; Sudek, Sebastian
- Molecular Biology and Evolution, Vol. 32, Issue 9
Cytoplasmic steps of peptidoglycan biosynthesis
journal, March 2008
- Barreteau, Hélène; Kovač, Andreja; Boniface, Audrey
- FEMS Microbiology Reviews, Vol. 32, Issue 2
Metagenomes of the Picoalga Bathycoccus from the Chile Coastal Upwelling
journal, June 2012
- Vaulot, Daniel; Lepère, Cécile; Toulza, Eve
- PLoS ONE, Vol. 7, Issue 6
Genome-wide mapping of alternative splicing in Arabidopsis thaliana
journal, October 2009
- Filichkin, S. A.; Priest, H. D.; Givan, S. A.
- Genome Research, Vol. 20, Issue 1
Comparison of aerobic and photosynthetic Rhodobacter sphaeroides 2.4.1 proteomes
journal, December 2006
- Callister, Stephen J.; Nicora, Carrie D.; Zeng, Xiaohua
- Journal of Microbiological Methods, Vol. 67, Issue 3
Fungal Genomics
book, May 2014
- Kuo, Alan; Bushnell, Brian; Grigoriev, Igor V.
- Fungi, Vol. 70, p. 1-52
Spectral Probabilities and Generating Functions of Tandem Mass Spectra: A Strike against Decoy Databases
journal, August 2008
- Kim, Sangtae; Gupta, Nitin; Pevzner, Pavel A.
- Journal of Proteome Research, Vol. 7, Issue 8
Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells
journal, April 2011
- Wang, Yuexi; Yang, Feng; Gritsenko, Marina A.
- PROTEOMICS, Vol. 11, Issue 10
High-Throughput RNA Sequencing of Pseudomonas-Infected Arabidopsis Reveals Hidden Transcriptome Complexity and Novel Splice Variants
journal, October 2013
- Howard, Brian E.; Hu, Qiwen; Babaoglu, Ahmet Can
- PLoS ONE, Vol. 8, Issue 10
Diversity of Picoplanktonic Prasinophytes Assessed by Direct Nuclear SSU rDNA Sequencing of Environmental Samples and Novel Isolates Retrieved from Oceanic and Coastal Marine Ecosystems
journal, June 2004
- Guillou, Laure; Eikrem, Wenche; Chrétiennot-Dinet, Marie-Josèphe
- Protist, Vol. 155, Issue 2
Peptidoglycan: a post-genomic analysis
journal, January 2012
- Cayrou, Caroline; Henrissat, Bernard; Gouret, Philippe
- BMC Microbiology, Vol. 12, Issue 1
The discoidin domain family revisited: New members from prokaryotes and a homology-based fold prediction
journal, July 1998
- Baumgartner, Stefan; Hofmann, Kay; Bucher, Philipp
- Protein Science, Vol. 7, Issue 7

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal