Need for Laboratory Ecosystems To Unravel the Structures and Functions of Soil Microbial Communities Mediated by Chemistry
Abstract
ABSTRACT The chemistry underpinning microbial interactions provides an integrative framework for linking the activities of individual microbes, microbial communities, plants, and their environments. Currently, we know very little about the functions of genes and metabolites within these communities because genome annotations and functions are derived from the minority of microbes that have been propagated in the laboratory. Yet the diversity, complexity, inaccessibility, and irreproducibility of native microbial consortia limit our ability to interpret chemical signaling and map metabolic networks. In this perspective, we contend that standardized laboratory ecosystems are needed to dissect the chemistry of soil microbiomes. We argue that dissemination and application of standardized laboratory ecosystems will be transformative for the field, much like how model organisms have played critical roles in advancing biochemistry and molecular and cellular biology. Community consensus on fabricated ecosystems (“EcoFABs”) along with protocols and data standards will integrate efforts and enable rapid improvements in our understanding of the biochemical ecology of microbial communities.
- Authors:
- Publication Date:
- Research Org.:
- Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States); Johns Hopkins Univ., Baltimore, MD (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC), Biological and Environmental Research (BER); USDOE Office of Energy Efficiency and Renewable Energy (EERE)
- OSTI Identifier:
- 1460472
- Alternate Identifier(s):
- OSTI ID: 1478345; OSTI ID: 1485593
- Grant/Contract Number:
- SC0012586; SC0012658; SC0018344; AC02-05CH11231; SC0014079
- Resource Type:
- Published Article
- Journal Name:
- mBio (Online)
- Additional Journal Information:
- Journal Name: mBio (Online) Journal Volume: 9 Journal Issue: 4; Journal ID: ISSN 2150-7511
- Publisher:
- American Society for Microbiology
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 54 ENVIRONMENTAL SCIENCES; 37 INORGANIC, ORGANIC, PHYSICAL, AND ANALYTICAL CHEMISTRY; chemistry of soil microbiomes; exometabolomics; laboratory ecosystems; metabolic networks; synthetic communities
Citation Formats
Zhalnina, Kateryna, Zengler, Karsten, Newman, Dianne, Northen, Trent R., and Davies, ed., Julian E. Need for Laboratory Ecosystems To Unravel the Structures and Functions of Soil Microbial Communities Mediated by Chemistry. United States: N. p., 2018.
Web. doi:10.1128/mBio.01175-18.
Zhalnina, Kateryna, Zengler, Karsten, Newman, Dianne, Northen, Trent R., & Davies, ed., Julian E. Need for Laboratory Ecosystems To Unravel the Structures and Functions of Soil Microbial Communities Mediated by Chemistry. United States. https://doi.org/10.1128/mBio.01175-18
Zhalnina, Kateryna, Zengler, Karsten, Newman, Dianne, Northen, Trent R., and Davies, ed., Julian E. Tue .
"Need for Laboratory Ecosystems To Unravel the Structures and Functions of Soil Microbial Communities Mediated by Chemistry". United States. https://doi.org/10.1128/mBio.01175-18.
@article{osti_1460472,
title = {Need for Laboratory Ecosystems To Unravel the Structures and Functions of Soil Microbial Communities Mediated by Chemistry},
author = {Zhalnina, Kateryna and Zengler, Karsten and Newman, Dianne and Northen, Trent R. and Davies, ed., Julian E.},
abstractNote = {ABSTRACT The chemistry underpinning microbial interactions provides an integrative framework for linking the activities of individual microbes, microbial communities, plants, and their environments. Currently, we know very little about the functions of genes and metabolites within these communities because genome annotations and functions are derived from the minority of microbes that have been propagated in the laboratory. Yet the diversity, complexity, inaccessibility, and irreproducibility of native microbial consortia limit our ability to interpret chemical signaling and map metabolic networks. In this perspective, we contend that standardized laboratory ecosystems are needed to dissect the chemistry of soil microbiomes. We argue that dissemination and application of standardized laboratory ecosystems will be transformative for the field, much like how model organisms have played critical roles in advancing biochemistry and molecular and cellular biology. Community consensus on fabricated ecosystems (“EcoFABs”) along with protocols and data standards will integrate efforts and enable rapid improvements in our understanding of the biochemical ecology of microbial communities.},
doi = {10.1128/mBio.01175-18},
journal = {mBio (Online)},
number = 4,
volume = 9,
place = {United States},
year = {Tue Jul 17 00:00:00 EDT 2018},
month = {Tue Jul 17 00:00:00 EDT 2018}
}
https://doi.org/10.1128/mBio.01175-18
Web of Science
Figures / Tables:
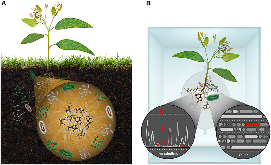 Figure 1: (A) Metabolites are the currency and communication for microbial communities. (B) Model laboratory ecosystems for discovering causal mechanisms of connections between genes and metabolites governing plant-microbe interactions.
Figure 1: (A) Metabolites are the currency and communication for microbial communities. (B) Model laboratory ecosystems for discovering causal mechanisms of connections between genes and metabolites governing plant-microbe interactions.
Works referenced in this record:
Networks of energetic and metabolic interactions define dynamics in microbial communities
journal, November 2015
- Embree, Mallory; Liu, Joanne K.; Al-Bassam, Mahmoud M.
- Proceedings of the National Academy of Sciences, Vol. 112, Issue 50
Feed Your Friends: Do Plant Exudates Shape the Root Microbiome?
journal, January 2018
- Sasse, Joelle; Martinoia, Enrico; Northen, Trent
- Trends in Plant Science, Vol. 23, Issue 1
Quantitative 3D Analysis of Plant Roots Growing in Soil Using Magnetic Resonance Imaging
journal, January 2016
- van Dusschoten, Dagmar; Metzner, Ralf; Kochs, Johannes
- Plant Physiology, Vol. 170, Issue 3
Large-scale de novo DNA synthesis: technologies and applications
journal, April 2014
- Kosuri, Sriram; Church, George M.
- Nature Methods, Vol. 11, Issue 5
GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems
journal, August 2015
- Rellán-Álvarez, Rubén; Lobet, Guillaume; Lindner, Heike
- eLife, Vol. 4
The RootChip: An Integrated Microfluidic Chip for Plant Science
journal, December 2011
- Grossmann, Guido; Guo, Woei-Jiun; Ehrhardt, David W.
- The Plant Cell, Vol. 23, Issue 12
Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases
journal, September 2014
- Citorik, Robert J.; Mimee, Mark; Lu, Timothy K.
- Nature Biotechnology, Vol. 32, Issue 11
Soil biodiversity and human health
journal, November 2015
- Wall, Diana H.; Nielsen, Uffe N.; Six, Johan
- Nature, Vol. 528, Issue 7580
Ecosystem Fabrication (EcoFAB) Protocols for The Construction of Laboratory Ecosystems Designed to Study Plant-microbe Interactions
journal, January 2018
- Gao, Jian; Sasse, Joelle; Lewald, Kyle M.
- Journal of Visualized Experiments, Issue 134
A metabolic pathway for catabolizing levulinic acid in bacteria
journal, September 2017
- Rand, Jacqueline M.; Pisithkul, Tippapha; Clark, Ryan L.
- Nature Microbiology, Vol. 2, Issue 12
Reinvigorating natural product combinatorial biosynthesis with synthetic biology
journal, August 2015
- Kim, Eunji; Moore, Bradley S.; Yoon, Yeo Joon
- Nature Chemical Biology, Vol. 11, Issue 9
Metabolic modeling of a mutualistic microbial community
journal, January 2007
- Stolyar, Sergey; Van Dien, Steve; Hillesland, Kristina Linnea
- Molecular Systems Biology, Vol. 3, Issue 1
Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla
journal, September 2012
- Wrighton, K. C.; Thomas, B. C.; Sharon, I.
- Science, Vol. 337, Issue 6102
Synergistic effects of soil microstructure and bacterial EPS on drying rate in emulated soil micromodels
journal, April 2015
- Deng, Jinzi; Orner, Erika P.; Chau, Jessica Furrer
- Soil Biology and Biochemistry, Vol. 83
Simplified and representative bacterial community of maize roots
journal, March 2017
- Niu, Ben; Paulson, Joseph Nathaniel; Zheng, Xiaoqi
- Proceedings of the National Academy of Sciences, Vol. 114, Issue 12
Heavy water and 15 N labelling with NanoSIMS analysis reveals growth rate-dependent metabolic heterogeneity in chemostats : NanoSIMS analysis of metabolic diversity in chemostats
journal, March 2015
- Kopf, Sebastian H.; McGlynn, Shawn E.; Green-Saxena, Abigail
- Environmental Microbiology, Vol. 17, Issue 7
New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters
journal, January 2016
- Luo, Yunzi; Enghiad, Behnam; Zhao, Huimin
- Natural Product Reports, Vol. 33, Issue 2
Pore-scale water dynamics during drying and the impacts of structure and surface wettability: PORE-SCALE WATER DYNAMICS DURING DRYING
journal, July 2017
- Cruz, Brian C.; Furrer, Jessica M.; Guo, Yi-Syuan
- Water Resources Research, Vol. 53, Issue 7
A road map for the development of community systems (CoSy) biology
journal, March 2012
- Zengler, Karsten; Palsson, Bernhard O.
- Nature Reviews Microbiology, Vol. 10, Issue 5
Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls
journal, November 2016
- Kallenbach, Cynthia M.; Frey, Serita D.; Grandy, A. Stuart
- Nature Communications, Vol. 7, Issue 1
Persistence of soil organic matter as an ecosystem property
journal, October 2011
- Schmidt, Michael W. I.; Torn, Margaret S.; Abiven, Samuel
- Nature, Vol. 478, Issue 7367
Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce
journal, April 2014
- Schreiter, Susanne; Ding, Guo-Chun; Heuer, Holger
- Frontiers in Microbiology, Vol. 5
Protein-SIP in environmental studies
journal, October 2016
- Jehmlich, Nico; Vogt, Carsten; Lünsmann, Vanessa
- Current Opinion in Biotechnology, Vol. 41
Multiscale imaging of plants: current approaches and challenges
journal, January 2015
- Rousseau, David; Chéné, Yann; Belin, Etienne
- Plant Methods, Vol. 11, Issue 1
Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli
journal, December 2016
- Sévin, Daniel C.; Fuhrer, Tobias; Zamboni, Nicola
- Nature Methods, Vol. 14, Issue 2
Constraint-based models predict metabolic and associated cellular functions
journal, January 2014
- Bordbar, Aarash; Monk, Jonathan M.; King, Zachary A.
- Nature Reviews Genetics, Vol. 15, Issue 2
Visualizing in situ translational activity for identifying and sorting slow-growing archaeal−bacterial consortia
journal, June 2016
- Hatzenpichler, Roland; Connon, Stephanie A.; Goudeau, Danielle
- Proceedings of the National Academy of Sciences, Vol. 113, Issue 28
Understanding soil food web dynamics, how close do we get?
journal, November 2016
- Morriën, E.
- Soil Biology and Biochemistry, Vol. 102
Signaling in the Rhizosphere
journal, March 2016
- Venturi, Vittorio; Keel, Christoph
- Trends in Plant Science, Vol. 21, Issue 3
A communal catalogue reveals Earth’s multiscale microbial diversity
journal, November 2017
- Thompson, Luke R.; Sanders, Jon G.; McDonald, Daniel
- Nature, Vol. 551, Issue 7681
Evidence-Based Annotation of Gene Function in Shewanella oneidensis MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
journal, November 2011
- Deutschbauer, Adam; Price, Morgan N.; Wetmore, Kelly M.
- PLoS Genetics, Vol. 7, Issue 11
The Colorful World of Extracellular Electron Shuttles
journal, September 2017
- Glasser, Nathaniel R.; Saunders, Scott H.; Newman, Dianne K.
- Annual Review of Microbiology, Vol. 71, Issue 1
The Future of Cell Biology: Emerging Model Organisms
journal, November 2016
- Goldstein, Bob; King, Nicole
- Trends in Cell Biology, Vol. 26, Issue 11
Dispersing misconceptions and identifying opportunities for the use of 'omics' in soil microbial ecology
journal, June 2015
- Prosser, James I.
- Nature Reviews Microbiology, Vol. 13, Issue 7
Discovery of MRSA active antibiotics using primary sequence from the human microbiome
journal, October 2016
- Chu, John; Vila-Farres, Xavier; Inoyama, Daigo
- Nature Chemical Biology, Vol. 12, Issue 12
Elucidation of complexity and prediction of interactions in microbial communities
journal, August 2017
- Zuñiga, Cristal; Zaramela, Livia; Zengler, Karsten
- Microbial Biotechnology, Vol. 10, Issue 6
Microbial contributions to climate change through carbon cycle feedbacks
journal, July 2008
- Bardgett, Richard D.; Freeman, Chris; Ostle, Nicholas J.
- The ISME Journal, Vol. 2, Issue 8
Toward a Predictive Understanding of Earth’s Microbiomes to Address 21st Century Challenges
journal, May 2016
- Blaser, Martin J.; Cardon, Zoe G.; Cho, Mildred K.
- mBio, Vol. 7, Issue 3
Biochemical Ecology of Soil Microorganisms
journal, October 1964
- Alexander, M.
- Annual Review of Microbiology, Vol. 18, Issue 1
Live imaging of root–bacteria interactions in a microfluidics setup
journal, March 2017
- Massalha, Hassan; Korenblum, Elisa; Malitsky, Sergey
- Proceedings of the National Academy of Sciences, Vol. 114, Issue 17
Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens
journal, February 2018
- Hover, Bradley M.; Kim, Seong-Hwan; Katz, Micah
- Nature Microbiology, Vol. 3, Issue 4
Using stable isotopes to explore root-microbe-mineral interactions in soil
journal, June 2017
- Pett-Ridge, Jennifer; Firestone, Mary K.
- Rhizosphere, Vol. 3
Enzymatic Degradation of Phenazines Can Generate Energy and Protect Sensitive Organisms from Toxicity
journal, October 2015
- Costa, Kyle C.; Bergkessel, Megan; Saunders, Scott
- mBio, Vol. 6, Issue 6
Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii
journal, January 2013
- Nagarajan, Harish; Sahin, Merve; Nogales, Juan
- Microbial Cell Factories, Vol. 12, Issue 1
Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist
journal, August 2015
- Cohen, Louis J.; Kang, Hahk-Soo; Chu, John
- Proceedings of the National Academy of Sciences, Vol. 112, Issue 35
A new view of the tree of life
journal, April 2016
- Hug, Laura A.; Baker, Brett J.; Anantharaman, Karthik
- Nature Microbiology, Vol. 1, Issue 5
Genomes from Metagenomics
journal, November 2013
- Sharon, I.; Banfield, J. F.
- Science, Vol. 342, Issue 6162
Redox Fluctuations Frame Microbial Community Impacts on N-cycling Rates in a Humid Tropical Forest Soil
journal, July 2006
- Pett-Ridge, Jennifer; Silver, Whendee L.; Firestone, Mary K.
- Biogeochemistry, Vol. 81, Issue 1
Tracking heavy water (D 2 O) incorporation for identifying and sorting active microbial cells
journal, December 2014
- Berry, David; Mader, Esther; Lee, Tae Kwon
- Proceedings of the National Academy of Sciences, Vol. 112, Issue 2
Culture-independent discovery of natural products from soil metagenomes
journal, November 2015
- Katz, Micah; Hover, Bradley M.; Brady, Sean F.
- Journal of Industrial Microbiology & Biotechnology, Vol. 43, Issue 2-3
Micro-scale determinants of bacterial diversity in soil
journal, November 2013
- Vos, Michiel; Wolf, Alexandra B.; Jennings, Sarah J.
- FEMS Microbiology Reviews, Vol. 37, Issue 6
Insights into Secondary Metabolism from a Global Analysis of Prokaryotic Biosynthetic Gene Clusters
journal, July 2014
- Cimermancic, Peter; Medema, Marnix H.; Claesen, Jan
- Cell, Vol. 158, Issue 2
Evolution of species interactions determines microbial community productivity in new environments
journal, November 2014
- Fiegna, Francesca; Moreno-Letelier, Alejandra; Bell, Thomas
- The ISME Journal, Vol. 9, Issue 5
Metabolic footprinting and systems biology: the medium is the message
journal, June 2005
- Kell, Douglas B.; Brown, Marie; Davey, Hazel M.
- Nature Reviews Microbiology, Vol. 3, Issue 7
Genome-wide identification of bacterial plant colonization genes
journal, September 2017
- Cole, Benjamin J.; Feltcher, Meghan E.; Waters, Robert J.
- PLOS Biology, Vol. 15, Issue 9
Scaling laws predict global microbial diversity
journal, May 2016
- Locey, Kenneth J.; Lennon, Jay T.
- Proceedings of the National Academy of Sciences, Vol. 113, Issue 21
Mining the Metabiome: Identifying Novel Natural Products from Microbial Communities
journal, September 2014
- Milshteyn, Aleksandr; Schneider, Jessica S.; Brady, Sean F.
- Chemistry & Biology, Vol. 21, Issue 9
A global atlas of the dominant bacteria found in soil
journal, January 2018
- Delgado-Baquerizo, Manuel; Oliverio, Angela M.; Brewer, Tess E.
- Science, Vol. 359, Issue 6373
Metabolomics for Secondary Metabolite Research
journal, November 2013
- Breitling, Rainer; Ceniceros, Ana; Jankevics, Andris
- Metabolites, Vol. 3, Issue 4
Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms
journal, March 2012
- Caporaso, J. Gregory; Lauber, Christian L.; Walters, William A.
- The ISME Journal, Vol. 6, Issue 8
Prokaryotic Diversity--Magnitude, Dynamics, and Controlling Factors
journal, May 2002
- Torsvik, V.
- Science, Vol. 296, Issue 5570
Root microbiota drive direct integration of phosphate stress and immunity
journal, March 2017
- Castrillo, Gabriel; Teixeira, Paulo José Pereira Lima; Paredes, Sur Herrera
- Nature, Vol. 543, Issue 7646
The Microbial Engines That Drive Earth's Biogeochemical Cycles
journal, May 2008
- Falkowski, P. G.; Fenchel, T.; Delong, E. F.
- Science, Vol. 320, Issue 5879
New CRISPR–Cas systems from uncultivated microbes
journal, December 2016
- Burstein, David; Harrington, Lucas B.; Strutt, Steven C.
- Nature, Vol. 542, Issue 7640
Metabolomics by numbers: acquiring and understanding global metabolite data
journal, May 2004
- Goodacre, Royston; Vaidyanathan, Seetharaman; Dunn, Warwick B.
- Trends in Biotechnology, Vol. 22, Issue 5
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal