Metagenomic insights into diazotrophic communities across Arctic glacier forefields
Abstract
Microbial nitrogen fixation is crucial for building labile nitrogen stocks and facilitating higher plant colonisation in oligotrophic glacier forefield soils. Here, the diazotrophic bacterial community structure across four Arctic glacier forefields was investigated using metagenomic analysis. In total, 70 soil metagenomes were used for taxonomic interpretation based on 185 nitrogenase (nif) sequences, extracted from assembled contigs. The low number of recovered genes highlights the need for deeper sequencing in some diverse samples, to uncover the complete microbial populations. A key group of forefield diazotrophs, found throughout the forefields, was identified using a nifH phylogeny, associated with nifH Cluster I and III. Sequences related most closely to groups including Alphaproteobacteria, Betaproteobacteria, Cyanobacteria and Firmicutes. Using multiple nif genes in a Last Common Ancestor analysis revealed a diverse range of diazotrophs across the forefields. Key organisms identified across the forefields included Nostoc, Geobacter, Polaromonas and Frankia. Nitrogen fixers that are symbiotic with plants were also identified, through the presence of root associated diazotrophs, which fix nitrogen in return for reduced carbon. Additional nitrogen fixers identified in forefield soils were metabolically diverse, including fermentative and sulphur cycling bacteria, halophiles and anaerobes.
- Authors:
-
- School of Geographical Sciences, University of Bristol, UK
- School of Life Sciences, University of Bristol, UK
- DOE Joint Genome Institute, 2800 Mitchell Drive, Walnut Creek, CA 94598, US
- GFZ German Research Centre for Geosciences, Telegrafenenberg, 14473 Potsdam, Germany, School of Earth and Environment, University of Leeds, LS2 9JT, Leeds, UK, Department of Earth Sciences, Free University of Berlin, Malteserstr, 74-100, Building A, 12249, Berlin, Germany
- Publication Date:
- Research Org.:
- Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States). National Energy Research Scientific Computing Center (NERSC); Univ. of California, Oakland, CA (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC)
- OSTI Identifier:
- 1461009
- Alternate Identifier(s):
- OSTI ID: 1543956
- Grant/Contract Number:
- AC02-05CH11231
- Resource Type:
- Published Article
- Journal Name:
- FEMS Microbiology Ecology (Online)
- Additional Journal Information:
- Journal Name: FEMS Microbiology Ecology (Online) Journal Volume: 94 Journal Issue: 9; Journal ID: ISSN 1574-6941
- Publisher:
- Federation of European Microbiological Societies
- Country of Publication:
- Netherlands
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES; Microbiology; nitrogen fixation; metagenomics; forefield; Arctic; diversity; diazotrophs
Citation Formats
Nash, Maisie V., Anesio, Alexandre M., Barker, Gary, Tranter, Martyn, Varliero, Gilda, Eloe-Fadrosh, Emiley A., Nielsen, Torben, Turpin-Jelfs, Thomas, Benning, Liane G., and Sánchez-Baracaldo, Patricia. Metagenomic insights into diazotrophic communities across Arctic glacier forefields. Netherlands: N. p., 2018.
Web. doi:10.1093/femsec/fiy114.
Nash, Maisie V., Anesio, Alexandre M., Barker, Gary, Tranter, Martyn, Varliero, Gilda, Eloe-Fadrosh, Emiley A., Nielsen, Torben, Turpin-Jelfs, Thomas, Benning, Liane G., & Sánchez-Baracaldo, Patricia. Metagenomic insights into diazotrophic communities across Arctic glacier forefields. Netherlands. https://doi.org/10.1093/femsec/fiy114
Nash, Maisie V., Anesio, Alexandre M., Barker, Gary, Tranter, Martyn, Varliero, Gilda, Eloe-Fadrosh, Emiley A., Nielsen, Torben, Turpin-Jelfs, Thomas, Benning, Liane G., and Sánchez-Baracaldo, Patricia. Tue .
"Metagenomic insights into diazotrophic communities across Arctic glacier forefields". Netherlands. https://doi.org/10.1093/femsec/fiy114.
@article{osti_1461009,
title = {Metagenomic insights into diazotrophic communities across Arctic glacier forefields},
author = {Nash, Maisie V. and Anesio, Alexandre M. and Barker, Gary and Tranter, Martyn and Varliero, Gilda and Eloe-Fadrosh, Emiley A. and Nielsen, Torben and Turpin-Jelfs, Thomas and Benning, Liane G. and Sánchez-Baracaldo, Patricia},
abstractNote = {Microbial nitrogen fixation is crucial for building labile nitrogen stocks and facilitating higher plant colonisation in oligotrophic glacier forefield soils. Here, the diazotrophic bacterial community structure across four Arctic glacier forefields was investigated using metagenomic analysis. In total, 70 soil metagenomes were used for taxonomic interpretation based on 185 nitrogenase (nif) sequences, extracted from assembled contigs. The low number of recovered genes highlights the need for deeper sequencing in some diverse samples, to uncover the complete microbial populations. A key group of forefield diazotrophs, found throughout the forefields, was identified using a nifH phylogeny, associated with nifH Cluster I and III. Sequences related most closely to groups including Alphaproteobacteria, Betaproteobacteria, Cyanobacteria and Firmicutes. Using multiple nif genes in a Last Common Ancestor analysis revealed a diverse range of diazotrophs across the forefields. Key organisms identified across the forefields included Nostoc, Geobacter, Polaromonas and Frankia. Nitrogen fixers that are symbiotic with plants were also identified, through the presence of root associated diazotrophs, which fix nitrogen in return for reduced carbon. Additional nitrogen fixers identified in forefield soils were metabolically diverse, including fermentative and sulphur cycling bacteria, halophiles and anaerobes.},
doi = {10.1093/femsec/fiy114},
journal = {FEMS Microbiology Ecology (Online)},
number = 9,
volume = 94,
place = {Netherlands},
year = {Tue Jun 12 00:00:00 EDT 2018},
month = {Tue Jun 12 00:00:00 EDT 2018}
}
https://doi.org/10.1093/femsec/fiy114
Web of Science
Figures / Tables:
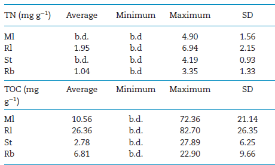 Table 1: Summary statistics for TN and TOC across the four fore-fields (Midtre Lovenbreen Ml, Russell Rl, Storglaciaren St and Rabots Rb). The average, minimum, maximum and standard deviation (SD) across each fore-field is given. The detection limit for both TN and TOC was 0.1 mg g−1. Sites recording valuesmore »
Table 1: Summary statistics for TN and TOC across the four fore-fields (Midtre Lovenbreen Ml, Russell Rl, Storglaciaren St and Rabots Rb). The average, minimum, maximum and standard deviation (SD) across each fore-field is given. The detection limit for both TN and TOC was 0.1 mg g−1. Sites recording valuesmore »
Works referenced in this record:
Thiocystis chemoclinalis sp. nov. and Thiocystis cadagnonensis sp. nov., motile purple sulfur bacteria isolated from the chemocline of a meromictic lake
journal, August 2010
- Peduzzi, S.; Welsh, A.; Demarta, A.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 61, Issue 7
Development and experimental validation of a nifH oligonucleotide microarray to study diazotrophic communities in a glacier forefield
journal, August 2009
- Duc, Laurence; Neuenschwander, Stefan; Rehrauer, Hubert
- Environmental Microbiology, Vol. 11, Issue 8
The role of microorganisms at different stages of ecosystem development for soil formation
journal, January 2013
- Schulz, S.; Brankatschk, R.; Dümig, A.
- Biogeosciences, Vol. 10, Issue 6
Complete genome sequence of Halorhodospira halophila SL1
journal, April 2013
- Challacombe, Jean F.; Majid, Sophia; Deole, Ratnakar
- Standards in Genomic Sciences, Vol. 8, Issue 2
Community assembly along proglacial chronosequences in the high Arctic: vegetation and soil development in north-west Svalbard
journal, August 2003
- Hodkinson, Ian D.; Coulson, Stephen J.; Webb, Nigel R.
- Journal of Ecology, Vol. 91, Issue 4
The earliest stages of ecosystem succession in high-elevation (5000 metres above sea level), recently deglaciated soils
journal, August 2008
- Schmidt, S. K.; Reed, Sasha C.; Nemergut, Diana R.
- Proceedings of the Royal Society B: Biological Sciences, Vol. 275, Issue 1653
Bacterial diversity in a glacier foreland of the high Arctic
journal, March 2010
- SchÜTte, Ursel M. E.; Abdo, Zaid; Foster, James
- Molecular Ecology, Vol. 19
A Comparison of rpoB and 16S rRNA as Markers in Pyrosequencing Studies of Bacterial Diversity
journal, February 2012
- Vos, Michiel; Quince, Christopher; Pijl, Agata S.
- PLoS ONE, Vol. 7, Issue 2
Commercial DNA extraction kits impact observed microbial community composition in permafrost samples
journal, October 2013
- Vishnivetskaya, Tatiana A.; Layton, Alice C.; Lau, Maggie C. Y.
- FEMS Microbiology Ecology, Vol. 87, Issue 1
Structural Basis of Biological Nitrogen Fixation
journal, January 1996
- Howard, James B.; Rees, Douglas C.
- Chemical Reviews, Vol. 96, Issue 7
Fast and accurate short read alignment with Burrows-Wheeler transform
journal, May 2009
- Li, H.; Durbin, R.
- Bioinformatics, Vol. 25, Issue 14
Soil sulphur speciation in two glacier forefield soil chronosequences assessed by S K -edge XANES spectroscopy : S speciation in glacier forefield soils by XANES
journal, February 2013
- Prietzel, J.; Wu, Y.; Dümig, A.
- European Journal of Soil Science, Vol. 64, Issue 2
Bacterial, Archaeal and Fungal Succession in the Forefield of a Receding Glacier
journal, December 2011
- Zumsteg, Anita; Luster, Jörg; Göransson, Hans
- Microbial Ecology, Vol. 63, Issue 3
Detection of Diazotrophy in the Acetylene-Fermenting Anaerobe Pelobacter sp. Strain SFB93
journal, June 2017
- Akob, Denise M.; Baesman, Shaun M.; Sutton, John M.
- Applied and Environmental Microbiology, Vol. 83, Issue 17
Bacterial community structure and function change in association with colonizer plants during early primary succession in a glacier forefield
journal, March 2012
- Knelman, Joseph E.; Legg, Teresa M.; O’Neill, Sean P.
- Soil Biology and Biochemistry, Vol. 46
Nitrogen fixation in different biogeochemical niches along a 120 000-year chronosequence in New Zealand
journal, August 2009
- Menge, Duncan N. L.; Hedin, Lars O.
- Ecology, Vol. 90, Issue 8
Effects of pioneering plants on microbial structures and functions in a glacier forefield
journal, May 2007
- Miniaci, Ciro; Bunge, Michael; Duc, Laurence
- Biology and Fertility of Soils, Vol. 44, Issue 2
MEGAN Community Edition - Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data
journal, June 2016
- Huson, Daniel H.; Beier, Sina; Flade, Isabell
- PLOS Computational Biology, Vol. 12, Issue 6
The Actinorhizal Symbiosis
journal, June 2000
- Wall, Luis Gabriel
- Journal of Plant Growth Regulation, Vol. 19, Issue 2
Primary succession of soil Crenarchaeota across a receding glacier foreland
journal, March 2005
- Nicol, Graeme W.; Tscherko, Dagmar; Embley, T. Martin
- Environmental Microbiology, Vol. 7, Issue 3
Genome of Geobacter sulfurreducens: Metal Reduction in Subsurface Environments
journal, December 2003
- Methe, B. A.; Nelson, K. E.; Eisen, J. A.
- Science, Vol. 302, Issue 5652, p. 1967-1969
Differential expression of nifH and anfH genes in Paenibacillus durus analysed by reverse transcriptase-PCR and denaturing gradient gel electrophoresis
journal, March 2008
- Teixeira, R. L. F.; von der Weid, I.; Seldin, L.
- Letters in Applied Microbiology, Vol. 46, Issue 3
Regulation and Genetics of Bacterial Nitrogen Fixation
journal, October 1975
- Brill, W. J.
- Annual Review of Microbiology, Vol. 29, Issue 1
Functional assignment of metagenomic data: challenges and applications
journal, July 2012
- Prakash, T.; Taylor, T. D.
- Briefings in Bioinformatics, Vol. 13, Issue 6
Characterization and Description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an Aryl-Halorespiring Facultative Anaerobic Myxobacterium
journal, February 2002
- Sanford, R. A.; Cole, J. R.; Tiedje, J. M.
- Applied and Environmental Microbiology, Vol. 68, Issue 2
Why are most rhizobia beneficial to their plant hosts, rather than parasitic?
journal, November 2004
- Denison, R.; Tobykiers, E.
- Microbes and Infection, Vol. 6, Issue 13
Nitrogen fixation on Arctic glaciers, Svalbard
journal, January 2011
- Telling, Jon; Anesio, Alexandre M.; Tranter, Martyn
- Journal of Geophysical Research, Vol. 116, Issue G3
Metagenomics: Application of Genomics to Uncultured Microorganisms
journal, December 2004
- Handelsman, J.
- Microbiology and Molecular Biology Reviews, Vol. 68, Issue 4
Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition
journal, December 2009
- Sattin, Sarah R.; Cleveland, Cory C.; Hood, Eran
- The Journal of Microbiology, Vol. 47, Issue 6
Identification of grass-associated and toluene-degrading diazotrophs, Azoarcus spp., by analyses of partial 16S ribosomal DNA sequences.
journal, January 1995
- Hurek, T.; Reinhold-Hurek, B.
- Applied and environmental microbiology, Vol. 61, Issue 6
The Natural History of Nitrogen Fixation
journal, March 2004
- Raymond, Jason; Siefert, Janet L.; Staples, Christopher R.
- Molecular Biology and Evolution, Vol. 21, Issue 3
Crucial Role of Extracellular Polysaccharides in Desiccation and Freezing Tolerance in the Terrestrial Cyanobacterium Nostoc commune
journal, November 2005
- Tamaru, Y.; Takani, Y.; Yoshida, T.
- Applied and Environmental Microbiology, Vol. 71, Issue 11
Bacterial and fungal community responses to reciprocal soil transfer along a temperature and soil moisture gradient in a glacier forefield
journal, June 2013
- Zumsteg, Anita; Bååth, Erland; Stierli, Beat
- Soil Biology and Biochemistry, Vol. 61
High Diversity of Diazotrophs in the Forefield of a Receding Alpine Glacier
journal, June 2008
- Duc, Laurence; Noll, Matthias; Meier, Brigitte E.
- Microbial Ecology, Vol. 57, Issue 1
Weathering-Associated Bacteria from the Damma Glacier Forefield: Physiological Capabilities and Impact on Granite Dissolution
journal, June 2010
- Frey, B.; Rieder, S. R.; Brunner, I.
- Applied and Environmental Microbiology, Vol. 76, Issue 14
Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism.
journal, January 1994
- Caccavo, F.; Lonergan, D. J.; Lovley, D. R.
- Applied and Environmental Microbiology, Vol. 60, Issue 10
Pioneering fungi from the Damma glacier forefield in the Swiss Alps can promote granite weathering: Pioneering fungi
journal, February 2011
- Brunner, I.; PlÖTze, M.; Rieder, S.
- Geobiology, Vol. 9, Issue 3
Arctic and subarctic soil populations of Rhizobium leguminosarum biovar trifolii nodulating three different clover species: Characterisation by diversity at chromosomal and symbiosis loci
journal, August 2005
- Fagerli, Inger Lise; Svenning, Mette M.
- Plant and Soil, Vol. 275, Issue 1-2
Metagenomic gene discovery: past, present and future
journal, June 2005
- Cowan, Don; Meyer, Quinton; Stafford, William
- Trends in Biotechnology, Vol. 23, Issue 6
Thioflavicoccus mobilis gen. nov., sp. nov., a novel purple sulfur bacterium with bacteriochlorophyll b.
journal, January 2001
- Imhoff, J. F.; Pfennig, N.
- International Journal of Systematic and Evolutionary Microbiology, Vol. 51, Issue 1
Nitroaromatic compounds serve as nitrogen source for Desulfovibrio sp. (B strain)
journal, April 1993
- Boopathy, R.; Kulpa, C. F.
- Canadian Journal of Microbiology, Vol. 39, Issue 4
Symbiotic Nitrogen Fixation
journal, July 1995
- Mylona, Panagiota; Pawlowski, Katharina; Bisseling, Ton
- The Plant Cell, Vol. 7, Issue 7
Variations in soil culturable bacteria communities and biochemical characteristics in the Dongkemadi glacier forefield along a chronosequence
journal, May 2012
- Liu, Guang-Xiu; Hu, Ping; Zhang, Wei
- Folia Microbiologica, Vol. 57, Issue 6
A Primer on Metagenomics
journal, February 2010
- Wooley, John C.; Godzik, Adam; Friedberg, Iddo
- PLoS Computational Biology, Vol. 6, Issue 2
Effect of DNA Extraction Method on the Apparent Microbial Diversity of Soil
journal, March 2010
- Inceoglu, O.; Hoogwout, E. F.; Hill, P.
- Applied and Environmental Microbiology, Vol. 76, Issue 10
Geobacter metallireducens accesses insoluble Fe(iii) oxide by chemotaxis
journal, April 2002
- Childers, Susan E.; Ciufo, Stacy; Lovley, Derek R.
- Nature, Vol. 416, Issue 6882
Changes in enzyme activities and soil microbial community composition along carbon and nutrient gradients at the Franz Josef chronosequence, New Zealand
journal, July 2007
- Allison, V. J.; Condron, L. M.; Peltzer, D. A.
- Soil Biology and Biochemistry, Vol. 39, Issue 7
Carbon and nitrogen determinations of carbonate-containing solids1
journal, May 1984
- Hedges, John I.; Stern, Jeffrey H.
- Limnology and Oceanography, Vol. 29, Issue 3
Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees
journal, April 2016
- He, Zilong; Zhang, Huangkai; Gao, Shenghan
- Nucleic Acids Research, Vol. 44, Issue W1
RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies
journal, January 2014
- Stamatakis, Alexandros
- Bioinformatics, Vol. 30, Issue 9
Microbial dynamics in glacier forefield soils show succession is not just skin deep
journal, February 2015
- Edwards, Arwyn; Cook, Sophie
- Molecular Ecology, Vol. 24, Issue 5
Hydrological controls on microbial communities in subglacial environments
journal, January 2005
- Tranter, Martyn; Skidmore, Mark; Wadham, Jemma
- Hydrological Processes, Vol. 19, Issue 4
Tackling soil diversity with the assembly of large, complex metagenomes
journal, March 2014
- Howe, Adina Chuang; Jansson, Janet K.; Malfatti, Stephanie A.
- Proceedings of the National Academy of Sciences, Vol. 111, Issue 13
Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield
journal, January 2015
- Rime, Thomas; Hartmann, Martin; Brunner, Ivano
- Molecular Ecology, Vol. 24, Issue 5
Comparison of different assembly and annotation tools on analysis of simulated viral metagenomic communities in the gut
journal, January 2014
- Vázquez-Castellanos, Jorge F.; García-López, Rodrigo; Pérez-Brocal, Vicente
- BMC Genomics, Vol. 15, Issue 1
Activation of the Jasmonic Acid Plant Defence Pathway Alters the Composition of Rhizosphere Bacterial Communities
journal, February 2013
- Carvalhais, Lilia C.; Dennis, Paul G.; Badri, Dayakar V.
- PLoS ONE, Vol. 8, Issue 2
Microbial metabolic networks in a complex electrogenic biofilm recovered from a stimulus-induced metatranscriptomics approach
journal, October 2015
- Ishii, Shun’ichi; Suzuki, Shino; Tenney, Aaron
- Scientific Reports, Vol. 5, Issue 1
Life in soil by the actinorhizal root nodule endophyte Frankia. A review
journal, September 2010
- Chaia, Eugenia E.; Wall, Luis G.; Huss-Danell, Kerstin
- Symbiosis, Vol. 51, Issue 3
SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing
journal, May 2012
- Bankevich, Anton; Nurk, Sergey; Antipov, Dmitry
- Journal of Computational Biology, Vol. 19, Issue 5
Functional Genomic Analysis of Three Nitrogenase Isozymes in the Photosynthetic Bacterium Rhodopseudomonas palustris
journal, November 2005
- Oda, Y.; Samanta, S. K.; Rey, F. E.
- Journal of Bacteriology, Vol. 187, Issue 22
Abundances and potential activities of nitrogen cycling microbial communities along a chronosequence of a glacier forefield
journal, December 2010
- Brankatschk, Robert; Töwe, Stefanie; Kleineidam, Kristina
- The ISME Journal, Vol. 5, Issue 6
The metagenomics of soil
journal, June 2005
- Daniel, Rolf
- Nature Reviews Microbiology, Vol. 3, Issue 6
VSEARCH: a versatile open source tool for metagenomics
journal, January 2016
- Rognes, Torbjørn; Flouri, Tomáš; Nichols, Ben
- PeerJ, Vol. 4
Geobacter uraniireducens sp. nov., isolated from subsurface sediment undergoing uranium bioremediation
journal, May 2008
- Shelobolina, E. S.; Vrionis, H. A.; Findlay, R. H.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 58, Issue 5
Shifts in desulfonating bacterial communities along a soil chronosequence in the forefield of a receding glacier
journal, February 2010
- Schmalenberger, Achim; Noll, Matthias
- FEMS Microbiology Ecology, Vol. 71, Issue 2
Molecular characterization of phototrophic microorganisms in the forefield of a receding glacier in the Swiss Alps
journal, March 2013
- Frey, Beat; Bühler, Lukas; Schmutz, Stefan
- Environmental Research Letters, Vol. 8, Issue 1
Microbial Assemblages in Soil Microbial Succession After Glacial Retreat in Svalbard (High Arctic)
journal, October 2005
- Kaštovská, Klára; Elster, Josef; Stibal, Marek
- Microbial Ecology, Vol. 50, Issue 3
SHIMMER (1.0): a novel mathematical model for microbial and biogeochemical dynamics in glacier forefield ecosystems
journal, January 2015
- Bradley, J. A.; Anesio, A. M.; Singarayer, J. S.
- Geoscientific Model Development Discussions, Vol. 8, Issue 8
Warming and drying suppress microbial activity and carbon cycling in boreal forest soils
journal, December 2008
- Allison, Steven D.; Treseder, Kathleen K.
- Global Change Biology, Vol. 14, Issue 12
Mechanism of bacterial adaptation to low temperature
journal, March 2006
- Chattopadhyay, M. K.
- Journal of Biosciences, Vol. 31, Issue 1
Metagenomics - a guide from sampling to data analysis
journal, February 2012
- Thomas, Torsten; Gilbert, Jack; Meyer, Folker
- Microbial Informatics and Experimentation, Vol. 2, Issue 1
Microbial dynamics in a High Arctic glacier forefield: a combined field,
laboratory, and modelling approach
journal, January 2016
- Bradley, James A.; Arndt, Sandra; Šabacká, Marie
- Biogeosciences, Vol. 13, Issue 19
Fermentative degradation of polyethylene glycol by a strictly anaerobic, gram-negative, nonsporeforming bacterium, Pelobacter venetianus sp. nov.
journal, January 1983
- Schink, B.; Stieb, M.
- Applied and Environmental Microbiology, Vol. 45, Issue 6
Genetic regulation of biological nitrogen fixation
journal, August 2004
- Dixon, Ray; Kahn, Daniel
- Nature Reviews Microbiology, Vol. 2, Issue 8
Molecular Diversity of nifH Genes from Bacteria Associated with High Arctic Dwarf Shrubs
journal, April 2006
- Deslippe, Julie R.; Egger, Keith N.
- Microbial Ecology, Vol. 51, Issue 4
SATé-II: Very Fast and Accurate Simultaneous Estimation of Multiple Sequence Alignments and Phylogenetic Trees
journal, December 2011
- Liu, Kevin; Warnow, Tandy J.; Holder, Mark T.
- Systematic Biology, Vol. 61, Issue 1
Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the archaeon Methanosarcina barkeri 227.
journal, January 1996
- Chien, Y. T.; Zinder, S. H.
- Journal of Bacteriology, Vol. 178, Issue 1
Nitrogenase gene diversity and microbial community structure: a cross-system comparison
journal, July 2003
- Zehr, Jonathan P.; Jenkins, Bethany D.; Short, Steven M.
- Environmental Microbiology, Vol. 5, Issue 7
Energy and Nitrogen Fixation
journal, September 1978
- Gutschick, Vincent P.
- BioScience, Vol. 28, Issue 9
The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes
journal, September 2008
- Meyer, F.; Paarmann, D.; D'Souza, M.
- BMC Bioinformatics, Vol. 9, Issue 1
IMG/M: integrated genome and metagenome comparative data analysis system
journal, October 2016
- Chen, I-Min A.; Markowitz, Victor M.; Chu, Ken
- Nucleic Acids Research, Vol. 45, Issue D1
Prokaryotic Diversity--Magnitude, Dynamics, and Controlling Factors
journal, May 2002
- Torsvik, V.
- Science, Vol. 296, Issue 5570
A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood
journal, October 2003
- Guindon, Stéphane; Gascuel, Olivier
- Systematic Biology, Vol. 52, Issue 5
Biology of Frankia strains, actinomycete symbionts of actinorhizal plants.
journal, January 1993
- Benson, D. R.; Silvester, W. B.
- Microbiological Reviews, Vol. 57, Issue 2
Chemical and Biological Gradients along the Damma Glacier Soil Chronosequence, Switzerland
journal, January 2011
- Bernasconi, Stefano M.; Bauder, Andreas; Bourdon, Bernard
- Vadose Zone Journal, Vol. 10, Issue 3
BFC: correcting Illumina sequencing errors
journal, May 2015
- Li, Heng
- Bioinformatics, Vol. 31, Issue 17
Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals
journal, April 1993
- Lovley, D. R.; Giovannoni, S. J.; White, D. C.
- Archives of Microbiology, Vol. 159, Issue 4
jModelTest 2: more models, new heuristics and parallel computing
journal, July 2012
- Darriba, Diego; Taboada, Guillermo L.; Doallo, Ramón
- Nature Methods, Vol. 9, Issue 8
Metagenomic and Small-Subunit rRNA Analyses Reveal the Genetic Diversity of Bacteria, Archaea, Fungi, and Viruses in Soil
journal, September 2007
- Fierer, N.; Breitbart, M.; Nulton, J.
- Applied and Environmental Microbiology, Vol. 73, Issue 21
Estimating coverage in metagenomic data sets and why it matters
journal, May 2014
- Rodriguez-R, Luis M.; Konstantinidis, Konstantinos T.
- The ISME Journal, Vol. 8, Issue 11
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal