Microbial Community Structure and Functional Potential Along a Hypersaline Gradient
Abstract
Salinity is one of the strongest environmental drivers of microbial evolution and community composition. Here we aimed to determine the impact of salt concentrations (2.5, 7.5, and 33.2%) on the microbial community structure of reclaimed saltern ponds near San Francisco, California, and to discover prospective enzymes with potential biotechnological applications. Community compositions were determined by 16S rRNA amplicon sequencing revealing both higher richness and evenness in the pond sediments compared to the water columns. Co-occurrence network analysis additionally uncovered the presence of microbial seed bank communities, potentially primed to respond to rapid changes in salinity. In addition, functional annotation of shotgun metagenomic DNA showed different capabilities if the microbial communities at different salinities for methanogenesis, amino acid metabolism, and carbohydrate-active enzymes. There was an overall shift with increasing salinity in the functional potential for starch degradation, and a decrease in degradation of cellulose and other oligosaccharides. Further, many carbohydrate-active enzymes identified have acidic isoelectric points that have potential biotechnological applications, including deconstruction of biofuel feedstocks under high ionic conditions. Metagenome-assembled genomes (MAGs) of individual halotolerant and halophilic microbes were binned revealing a variety of carbohydrate-degrading potential of individual pond inhabitants.
- Authors:
- Publication Date:
- Research Org.:
- Pacific Northwest National Laboratory (PNNL), Richland, WA (United States); Oak Ridge National Laboratory (ORNL), Oak Ridge, TN (United States)
- Sponsoring Org.:
- USDOE
- OSTI Identifier:
- 1459672
- Alternate Identifier(s):
- OSTI ID: 1461649; OSTI ID: 1471857
- Report Number(s):
- PNNL-SA-126820
Journal ID: ISSN 1664-302X; 1492
- Grant/Contract Number:
- AC02-05CH11231; AC05-76RL01830; AC05-00OR22725
- Resource Type:
- Published Article
- Journal Name:
- Frontiers in Microbiology
- Additional Journal Information:
- Journal Name: Frontiers in Microbiology Journal Volume: 9; Journal ID: ISSN 1664-302X
- Publisher:
- Frontiers Research Foundation
- Country of Publication:
- Switzerland
- Language:
- English
- Subject:
- 59 BASIC BIOLOGICAL SCIENCES; microbial communities; halophiles; biofuels; metagenomes; 16S rRNA
Citation Formats
Kimbrel, Jeffrey A., Ballor, Nicholas, Wu, Yu-Wei, David, Maude M., Hazen, Terry C., Simmons, Blake A., Singer, Steven W., and Jansson, Janet K. Microbial Community Structure and Functional Potential Along a Hypersaline Gradient. Switzerland: N. p., 2018.
Web. doi:10.3389/fmicb.2018.01492.
Kimbrel, Jeffrey A., Ballor, Nicholas, Wu, Yu-Wei, David, Maude M., Hazen, Terry C., Simmons, Blake A., Singer, Steven W., & Jansson, Janet K. Microbial Community Structure and Functional Potential Along a Hypersaline Gradient. Switzerland. https://doi.org/10.3389/fmicb.2018.01492
Kimbrel, Jeffrey A., Ballor, Nicholas, Wu, Yu-Wei, David, Maude M., Hazen, Terry C., Simmons, Blake A., Singer, Steven W., and Jansson, Janet K. Tue .
"Microbial Community Structure and Functional Potential Along a Hypersaline Gradient". Switzerland. https://doi.org/10.3389/fmicb.2018.01492.
@article{osti_1459672,
title = {Microbial Community Structure and Functional Potential Along a Hypersaline Gradient},
author = {Kimbrel, Jeffrey A. and Ballor, Nicholas and Wu, Yu-Wei and David, Maude M. and Hazen, Terry C. and Simmons, Blake A. and Singer, Steven W. and Jansson, Janet K.},
abstractNote = {Salinity is one of the strongest environmental drivers of microbial evolution and community composition. Here we aimed to determine the impact of salt concentrations (2.5, 7.5, and 33.2%) on the microbial community structure of reclaimed saltern ponds near San Francisco, California, and to discover prospective enzymes with potential biotechnological applications. Community compositions were determined by 16S rRNA amplicon sequencing revealing both higher richness and evenness in the pond sediments compared to the water columns. Co-occurrence network analysis additionally uncovered the presence of microbial seed bank communities, potentially primed to respond to rapid changes in salinity. In addition, functional annotation of shotgun metagenomic DNA showed different capabilities if the microbial communities at different salinities for methanogenesis, amino acid metabolism, and carbohydrate-active enzymes. There was an overall shift with increasing salinity in the functional potential for starch degradation, and a decrease in degradation of cellulose and other oligosaccharides. Further, many carbohydrate-active enzymes identified have acidic isoelectric points that have potential biotechnological applications, including deconstruction of biofuel feedstocks under high ionic conditions. Metagenome-assembled genomes (MAGs) of individual halotolerant and halophilic microbes were binned revealing a variety of carbohydrate-degrading potential of individual pond inhabitants.},
doi = {10.3389/fmicb.2018.01492},
journal = {Frontiers in Microbiology},
number = ,
volume = 9,
place = {Switzerland},
year = {Tue Jul 10 00:00:00 EDT 2018},
month = {Tue Jul 10 00:00:00 EDT 2018}
}
https://doi.org/10.3389/fmicb.2018.01492
Web of Science
Figures / Tables:
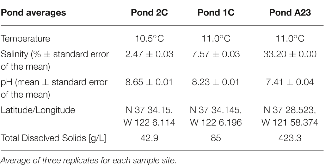 Table 1: Chemical analysis of pond water and sediments.
Table 1: Chemical analysis of pond water and sediments.
Works referenced in this record:
Microbial interactions: from networks to models
journal, July 2012
- Faust, Karoline; Raes, Jeroen
- Nature Reviews Microbiology, Vol. 10, Issue 8
Global patterns in bacterial diversity
journal, June 2007
- Lozupone, C. A.; Knight, R.
- Proceedings of the National Academy of Sciences, Vol. 104, Issue 27
Comparative genomics of the marine bacterial genus G laciecola reveals the high degree of genomic diversity and genomic characteristic for cold adaptation : Comparative genomics of the marine genus
journal, November 2013
- Qin, Qi-Long; Xie, Bin-Bin; Yu, Yong
- Environmental Microbiology, Vol. 16, Issue 6
Identification of GH15 Family Thermophilic Archaeal Trehalases That Function within a Narrow Acidic-pH Range
journal, May 2015
- Sakaguchi, Masayoshi; Shimodaira, Satoru; Ishida, Shin-nosuke
- Applied and Environmental Microbiology, Vol. 81, Issue 15
Genome Sequence of Halomonas sp. Strain KO116, an Ionic Liquid-Tolerant Marine Bacterium Isolated from a Lignin-Enriched Seawater Microcosm
journal, May 2015
- O’Dell, Kaela B.; Woo, Hannah L.; Utturkar, Sagar
- Genome Announcements, Vol. 3, Issue 3
Characterization of a metagenome-derived halotolerant cellulase
journal, October 2006
- Voget, S.; Steele, H. L.; Streit, W. R.
- Journal of Biotechnology, Vol. 126, Issue 1, p. 26-36
New Abundant Microbial Groups in Aquatic Hypersaline Environments
journal, October 2011
- Ghai, Rohit; Pašić, Lejla; Fernández, Ana Beatriz
- Scientific Reports, Vol. 1, Issue 1
Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus
journal, November 1999
- Ferreira, A. C.; Nobre, M. F.; Moore, E.
- Extremophiles, Vol. 3, Issue 4
Bioenergetic Aspects of Halophilism
journal, June 1999
- Oren, Aharon
- Microbiology and Molecular Biology Reviews, Vol. 63, Issue 2
Bacterial chitin utilisation at extremely haloalkaline conditions
journal, September 2012
- Sorokin, D. Y.; Tourova, T. P.; Sukhacheva, M. V.
- Extremophiles, Vol. 16, Issue 6
The ade4 Package: Implementing the Duality Diagram for Ecologists
journal, January 2007
- Dray, Stéphane; Dufour, Anne-Béatrice
- Journal of Statistical Software, Vol. 22, Issue 4
Glycosyltransferases and oligosaccharyltransferases in Archaea: putative components of the N -glycosylation pathway in the third domain of life
journal, November 2009
- Magidovich, Hilla; Eichler, Jerry
- FEMS Microbiology Letters, Vol. 300, Issue 1
Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea
journal, August 2017
- Bowers, Robert M.; Kyrpides, Nikos C.; Stepanauskas, Ramunas
- Nature Biotechnology, Vol. 35, Issue 8
ChloroplastDB: the Chloroplast Genome Database
journal, January 2006
- Cui, L.
- Nucleic Acids Research, Vol. 34, Issue 90001
Thiohalobacter thiocyanaticus gen. nov., sp. nov., a moderately halophilic, sulfur-oxidizing gammaproteobacterium from hypersaline lakes, that utilizes thiocyanate
journal, August 2009
- Sorokin, D. Yu.; Kovaleva, O. L.; Tourova, T. P.
- INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, Vol. 60, Issue 2
Phylogenetic Distribution of Potential Cellulases in Bacteria
journal, December 2012
- Berlemont, Renaud; Martiny, Adam C.
- Applied and Environmental Microbiology, Vol. 79, Issue 5
Fast and accurate short read alignment with Burrows-Wheeler transform
journal, May 2009
- Li, H.; Durbin, R.
- Bioinformatics, Vol. 25, Issue 14
Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids
journal, January 2009
- Pottkämper, Julia; Barthen, Peter; Ilmberger, Nele
- Green Chemistry, Vol. 11, Issue 7
Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications
journal, January 2002
- Oren, A.
- Journal of Industrial Microbiology & Biotechnology, Vol. 28, Issue 1
Spatial scales of bacterial community diversity at cold seeps (Eastern Mediterranean Sea)
journal, December 2014
- Pop Ristova, Petra; Wenzhöfer, Frank; Ramette, Alban
- The ISME Journal, Vol. 9, Issue 6
Data, information, knowledge and principle: back to metabolism in KEGG
journal, November 2013
- Kanehisa, Minoru; Goto, Susumu; Sato, Yoko
- Nucleic Acids Research, Vol. 42, Issue D1
Demonstrating microbial co-occurrence pattern analyses within and between ecosystems
journal, July 2014
- Williams, Ryan J.; Howe, Adina; Hofmockel, Kirsten S.
- Frontiers in Microbiology, Vol. 5
Comparison of dilute acid and ionic liquid pretreatment of switchgrass: Biomass recalcitrance, delignification and enzymatic saccharification
journal, July 2010
- Li, Chenlin; Knierim, Bernhard; Manisseri, Chithra
- Bioresource Technology, Vol. 101, Issue 13
Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community
journal, December 2013
- Podell, Sheila; Emerson, Joanne B.; Jones, Claudia M.
- The ISME Journal, Vol. 8, Issue 5
Genomic analysis reveals the biotechnological and industrial potential of levan producing halophilic extremophile, Halomonas smyrnensis AAD6T
journal, August 2015
- Diken, Elif; Ozer, Tugba; Arikan, Muzaffer
- SpringerPlus, Vol. 4, Issue 1
Biodegradation of Organic Pollutants by Halophilic Bacteria and Archaea
journal, January 2008
- Le Borgne, Sylvie; Paniagua, Dayanira; Vazquez-Duhalt, Rafael
- Journal of Molecular Microbiology and Biotechnology, Vol. 15, Issue 2-3
Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy
journal, June 2007
- Wang, Q.; Garrity, G. M.; Tiedje, J. M.
- Applied and Environmental Microbiology, Vol. 73, Issue 16
Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes
journal, January 2013
- Oren, Aharon
- Frontiers in Microbiology, Vol. 4
Prokaryotic Community Diversity Along an Increasing Salt Gradient in a Soda Ash Concentration Pond
journal, September 2015
- Simachew, Addis; Lanzén, Anders; Gessesse, Amare
- Microbial Ecology, Vol. 71, Issue 2
Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems
journal, March 2012
- Andrei, Adrian-Ştefan; Banciu, Horia Leonard; Oren, Aharon
- FEMS Microbiology Letters, Vol. 330, Issue 1
Gene cloning, expression and characterization of a new cold-active and salt-tolerant endo-β-1,4-xylanase from marine Glaciecola mesophila KMM 241
journal, June 2009
- Guo, Bing; Chen, Xiu-Lan; Sun, Cai-Yun
- Applied Microbiology and Biotechnology, Vol. 84, Issue 6
Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern
journal, June 2002
- Benlloch, Susana; Lopez-Lopez, Arantxa; Casamayor, Emilio O.
- Environmental Microbiology, Vol. 4, Issue 6
Identification of a haloalkaliphilic and thermostable cellulase with improved ionic liquid tolerance
journal, January 2011
- Zhang, Tao; Datta, Supratim; Eichler, Jerry
- Green Chemistry, Vol. 13, Issue 8
Prodigal: prokaryotic gene recognition and translation initiation site identification
journal, March 2010
- Hyatt, Doug; Chen, Gwo-Liang; LoCascio, Philip F.
- BMC Bioinformatics, Vol. 11, Issue 1
Relationship of sequence and structure to specificity in the α-amylase family of enzymes
journal, March 2001
- MacGregor, E. Ann; Janeček, Štefan; Svensson, Birte
- Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology, Vol. 1546, Issue 1
Prokaryotic taxonomic and metabolic diversity of an intermediate salinity hypersaline habitat assessed by metagenomics
journal, April 2014
- Fernández, Ana B.; Ghai, Rohit; Martin-Cuadrado, Ana-Belen
- FEMS Microbiology Ecology, Vol. 88, Issue 3
dbCAN: a web resource for automated carbohydrate-active enzyme annotation
journal, May 2012
- Yin, Yanbin; Mao, Xizeng; Yang, Jincai
- Nucleic Acids Research, Vol. 40, Issue W1
Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190
journal, March 2004
- Newberry, Carole J.; Webster, Gordon; Cragg, Barry A.
- Environmental Microbiology, Vol. 6, Issue 3
MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets
journal, October 2015
- Wu, Yu-Wei; Simmons, Blake A.; Singer, Steven W.
- Bioinformatics, Vol. 32, Issue 4
Cyanobacterial reuse of extracellular organic carbon in microbial mats
journal, October 2015
- Stuart, Rhona K.; Mayali, Xavier; Lee, Jackson Z.
- The ISME Journal, Vol. 10, Issue 5
CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes
journal, May 2015
- Parks, Donovan H.; Imelfort, Michael; Skennerton, Connor T.
- Genome Research, Vol. 25, Issue 7
Potential halophilic cellulases for in situ enzymatic saccharification of ionic liquids pretreated lignocelluloses
journal, March 2014
- Gunny, Ahmad Anas Nagoor; Arbain, Dachyar; Edwin Gumba, Rizo
- Bioresource Technology, Vol. 155
Primer and platform effects on 16S rRNA tag sequencing
journal, August 2015
- Tremblay, Julien; Singh, Kanwar; Fern, Alison
- Frontiers in Microbiology, Vol. 6
Molecular mechanisms underlying roseobacter–phytoplankton symbioses
journal, June 2010
- Geng, Haifeng; Belas, Robert
- Current Opinion in Biotechnology, Vol. 21, Issue 3
Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments
journal, September 1998
- Kempf, Bettina; Bremer, E.
- Archives of Microbiology, Vol. 170, Issue 5
Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture.
journal, January 1996
- González, J. M.; Whitman, W. B.; Hodson, R. E.
- Applied and environmental microbiology, Vol. 62, Issue 12
Extremophiles as a source for novel enzymes
journal, June 2003
- van den Burg, Bertus
- Current Opinion in Microbiology, Vol. 6, Issue 3
phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data
journal, April 2013
- McMurdie, Paul J.; Holmes, Susan
- PLoS ONE, Vol. 8, Issue 4
Light Regimes Shape Utilization of Extracellular Organic C and N in a Cyanobacterial Biofilm
journal, June 2016
- Stuart, Rhona K.; Mayali, Xavier; Boaro, Amy A.
- mBio, Vol. 7, Issue 3
Salt-activated endoglucanase of a strain of alkaliphilic Bacillus agaradhaerens
journal, April 2006
- Hirasawa, Kazumichi; Uchimura, Kohsuke; Kashiwa, Masami
- Antonie van Leeuwenhoek, Vol. 89, Issue 2
IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth
journal, April 2012
- Peng, Y.; Leung, H. C. M.; Yiu, S. M.
- Bioinformatics, Vol. 28, Issue 11
Recent trends in ionic liquid (IL) tolerant enzymes and microorganisms for biomass conversion
journal, October 2013
- Portillo, Maria del Carmen; Saadeddin, Anas
- Critical Reviews in Biotechnology, Vol. 35, Issue 3
Systematic Identification of Gene Families for Use as “Markers” for Phylogenetic and Phylogeny-Driven Ecological Studies of Bacteria and Archaea and Their Major Subgroups
journal, October 2013
- Wu, Dongying; Jospin, Guillaume; Eisen, Jonathan A.
- PLoS ONE, Vol. 8, Issue 10
How to be moderately halophilic with broad salt tolerance: clues from the genome of Chromohalobacter salexigens
journal, May 2005
- Oren, Aharon; Larimer, Frank; Richardson, Paul
- Extremophiles, Vol. 9, Issue 4
Complete Genome Sequence of the Marine Cellulose- and Xylan-Degrading Bacterium Glaciecolasp. Strain 4H-3-7+YE-5
journal, June 2011
- Klippel, B.; Lochner, A.; Bruce, D. C.
- Journal of Bacteriology, Vol. 193, Issue 17
Microbial Diversity in Deep-sea Methane Seep Sediments Presented by SSU rRNA Gene Tag Sequencing
journal, January 2012
- Nunoura, Takuro; Takaki, Yoshihiro; Kazama, Hiromi
- Microbes and environments, Vol. 27, Issue 4
Trehalose/2-sulfotrehalose biosynthesis and glycine-betaine uptake are widely spread mechanisms for osmoadaptation in the Halobacteriales
journal, September 2013
- Youssef, Noha H.; Savage-Ashlock, Kristen N.; McCully, Alexandra L.
- The ISME Journal, Vol. 8, Issue 3
The carbohydrate-active enzymes database (CAZy) in 2013
journal, November 2013
- Lombard, Vincent; Golaconda Ramulu, Hemalatha; Drula, Elodie
- Nucleic Acids Research, Vol. 42, Issue D1
Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes
journal, May 2013
- Albertsen, Mads; Hugenholtz, Philip; Skarshewski, Adam
- Nature Biotechnology, Vol. 31, Issue 6
Ecological Patterns Among Bacteria and Microbial Eukaryotes Derived from Network Analyses in a Low-Salinity Lake
journal, November 2017
- Jones, Adriane Clark; Hambright, K. David; Caron, David A.
- Microbial Ecology, Vol. 75, Issue 4
Using network analysis to explore co-occurrence patterns in soil microbial communities
journal, September 2011
- Barberán, Albert; Bates, Scott T.; Casamayor, Emilio O.
- The ISME Journal, Vol. 6, Issue 2
Biology of Moderately Halophilic Aerobic Bacteria
journal, June 1998
- Ventosa, Antonio; Nieto, Joaquín J.; Oren, Aharon
- Microbiology and Molecular Biology Reviews, Vol. 62, Issue 2
Halophilic hydrolases as a new tool for the biotechnological industries
journal, August 2012
- Delgado-García, Mariana; Valdivia-Urdiales, Blanca; Aguilar-González, Cristóbal Noe
- Journal of the Science of Food and Agriculture, Vol. 92, Issue 13
Variability in habitat value of commercial salt production ponds: implications for waterbird management and tidal marsh restoration planning
journal, May 2012
- Athearn, Nicole D.; Takekawa, John Y.; Bluso-Demers, Jill D.
- Hydrobiologia, Vol. 697, Issue 1
Genomic Potential for Polysaccharide Deconstruction in Bacteria
journal, December 2014
- Berlemont, Renaud; Martiny, Adam C.
- Applied and Environmental Microbiology, Vol. 81, Issue 4
BLAST+: architecture and applications
journal, January 2009
- Camacho, Christiam; Coulouris, George; Avagyan, Vahram
- BMC Bioinformatics, Vol. 10, Issue 1
Deciphering microbial interactions and detecting keystone species with co-occurrence networks
journal, May 2014
- Berry, David; Widder, Stefanie
- Frontiers in Microbiology, Vol. 5
Microbial diversity of hypersaline environments: a metagenomic approach
journal, June 2015
- Ventosa, Antonio; de la Haba, Rafael R.; Sánchez-Porro, Cristina
- Current Opinion in Microbiology, Vol. 25
Genome-wide analysis of lysine catabolism in bacteria reveals new connections with osmotic stress resistance
journal, July 2013
- Neshich, Izabella AP; Kiyota, Eduardo; Arruda, Paulo
- The ISME Journal, Vol. 7, Issue 12
Microbial seed banks: the ecological and evolutionary implications of dormancy
journal, January 2011
- Lennon, Jay T.; Jones, Stuart E.
- Nature Reviews Microbiology, Vol. 9, Issue 2
Xyloglucan Is Recognized by Carbohydrate-binding Modules That Interact with β-Glucan Chains
journal, November 2005
- Najmudin, Shabir; Guerreiro, Catarina I. P. D.; Carvalho, Ana L.
- Journal of Biological Chemistry, Vol. 281, Issue 13
Integrative analysis of environmental sequences using MEGAN4
journal, June 2011
- Huson, D. H.; Mitra, S.; Ruscheweyh, H. -J.
- Genome Research, Vol. 21, Issue 9
Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments
journal, April 2014
- Fathepure, Babu Z.
- Frontiers in Microbiology, Vol. 5
Sialic Acid Recognition by Vibrio cholerae Neuraminidase
journal, June 2004
- Moustafa, Ibrahim; Connaris, Helen; Taylor, Margaret
- Journal of Biological Chemistry, Vol. 279, Issue 39
FOAM (Functional Ontology Assignments for Metagenomes): a Hidden Markov Model (HMM) database with environmental focus
journal, September 2014
- Prestat, Emmanuel; David, Maude M.; Hultman, Jenni
- Nucleic Acids Research, Vol. 42, Issue 19
A novel salt-tolerant endo-β-1,4-glucanase Cel5A in Vibrio sp. G21 isolated from mangrove soil
journal, April 2010
- Gao, Zhaoming; Ruan, Lingwei; Chen, Xiulan
- Applied Microbiology and Biotechnology, Vol. 87, Issue 4, p. 1373-1382
Genome-Based Taxonomic Classification of Bacteroidetes
journal, December 2016
- Hahnke, Richard L.; Meier-Kolthoff, Jan P.; García-López, Marina
- Frontiers in Microbiology, Vol. 7
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
journal, December 2014
- Love, Michael I.; Huber, Wolfgang; Anders, Simon
- Genome Biology, Vol. 15, Issue 12
Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill
journal, January 2014
- Mason, Olivia U.; Scott, Nicole M.; Gonzalez, Antonio
- The ISME Journal, Vol. 8, Issue 7
Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes
journal, January 2013
- Levasseur, Anthony; Drula, Elodie; Lombard, Vincent
- Biotechnology for Biofuels, Vol. 6, Issue 1, Article No. 41

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal