Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei
Abstract
Microbial syntrophy is a vital metabolic interaction necessary for the complete oxidation of organic biomass to methane in all-anaerobic ecosystems. However, this process is thermodynamically constrained and represents an ecosystem-level metabolic bottleneck. To gain insight into the physiology of this process, a shotgun proteomics approach was used to quantify the protein landscape of the model syntrophic metabolizer, Syntrophomonas wolfei, grown axenically and syntrophically with Methanospirillum hungatei. Remarkably, the abundance of most proteins as represented by normalized spectral abundance factor (NSAF) value changed very little between the pure and coculture growth conditions. Among the most abundant proteins detected were GroEL and GroES chaperonins, a small heat shock protein, and proteins involved in electron transfer, beta-oxidation, and ATP synthesis. Several putative energy conservation enzyme systems that utilize NADH and ferredoxin were present. The abundance of an EtfAB2 and the membrane-bound iron-sulfur oxidoreductase (Swol_0698 gene product) delineated a potential conduit for electron transfer between acyl-CoA dehydrogenases and membrane redox carriers. Proteins detected only when S. wolfei was grown with M. hungatei included a zinc-dependent dehydrogenase with a GroES domain, whose gene is present in genomes in many organisms capable of syntrophy, and transcriptional regulators responsive to environmental stimuli or the physiological status ofmore »
- Authors:
-
- Univ. of Oklahoma, Norman, OK (United States). Dept. of Botany and Microbiology
- Univ. of Michigan, Ann Arbor, MI (United States)
- Oak Ridge National Lab. (ORNL), Oak Ridge, TN (United States). Chemical Sciences Division
- Univ. of California, Los Angeles, CA (United States)
- Publication Date:
- Research Org.:
- Univ. of California, Los Angeles, CA (United States); Univ. of Oklahoma, Norman, OK (United States); Oak Ridge National Laboratory (ORNL), Oak Ridge, TN (United States)
- Sponsoring Org.:
- USDOE Office of Science (SC), Basic Energy Sciences (BES). Chemical Sciences, Geosciences, and Biosciences Division; USDOE Office of Science (SC), Biological and Environmental Research (BER)
- Contributing Org.:
- Univ. of Oklahoma, Norman, OK (United States); Univ. of California, Los Angeles, CA (United States)
- OSTI Identifier:
- 1254004
- Alternate Identifier(s):
- OSTI ID: 1286726
- Grant/Contract Number:
- FC02-02ER63421; FG03-86ER13498; FG02-96ER20214; AC05-00OR22725
- Resource Type:
- Accepted Manuscript
- Journal Name:
- Frontiers in Microbiology
- Additional Journal Information:
- Journal Volume: 6; Journal ID: ISSN 1664-302X
- Publisher:
- Frontiers Research Foundation
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 60 APPLIED LIFE SCIENCES; 59 BASIC BIOLOGICAL SCIENCES; syntrophy; Syntrophomonas wolfei; interspecies electron transfer; reverse electron transfer; hydrogen; methanogenesis
Citation Formats
Sieber, Jessica R., Crable, Bryan R., Sheik, Cody S., Hurst, Gregory B., Rohlin, Lars, Gunsalus, Robert P., and McInerney, Michael J. Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei. United States: N. p., 2015.
Web. doi:10.3389/fmicb.2015.00115.
Sieber, Jessica R., Crable, Bryan R., Sheik, Cody S., Hurst, Gregory B., Rohlin, Lars, Gunsalus, Robert P., & McInerney, Michael J. Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei. United States. https://doi.org/10.3389/fmicb.2015.00115
Sieber, Jessica R., Crable, Bryan R., Sheik, Cody S., Hurst, Gregory B., Rohlin, Lars, Gunsalus, Robert P., and McInerney, Michael J. Wed .
"Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei". United States. https://doi.org/10.3389/fmicb.2015.00115. https://www.osti.gov/servlets/purl/1254004.
@article{osti_1254004,
title = {Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei},
author = {Sieber, Jessica R. and Crable, Bryan R. and Sheik, Cody S. and Hurst, Gregory B. and Rohlin, Lars and Gunsalus, Robert P. and McInerney, Michael J.},
abstractNote = {Microbial syntrophy is a vital metabolic interaction necessary for the complete oxidation of organic biomass to methane in all-anaerobic ecosystems. However, this process is thermodynamically constrained and represents an ecosystem-level metabolic bottleneck. To gain insight into the physiology of this process, a shotgun proteomics approach was used to quantify the protein landscape of the model syntrophic metabolizer, Syntrophomonas wolfei, grown axenically and syntrophically with Methanospirillum hungatei. Remarkably, the abundance of most proteins as represented by normalized spectral abundance factor (NSAF) value changed very little between the pure and coculture growth conditions. Among the most abundant proteins detected were GroEL and GroES chaperonins, a small heat shock protein, and proteins involved in electron transfer, beta-oxidation, and ATP synthesis. Several putative energy conservation enzyme systems that utilize NADH and ferredoxin were present. The abundance of an EtfAB2 and the membrane-bound iron-sulfur oxidoreductase (Swol_0698 gene product) delineated a potential conduit for electron transfer between acyl-CoA dehydrogenases and membrane redox carriers. Proteins detected only when S. wolfei was grown with M. hungatei included a zinc-dependent dehydrogenase with a GroES domain, whose gene is present in genomes in many organisms capable of syntrophy, and transcriptional regulators responsive to environmental stimuli or the physiological status of the cell. The proteomic analysis revealed an emphasis on macromolecular stability and energy metabolism by S. wolfei and presence of regulatory mechanisms responsive to external stimuli and cellular physiological status.},
doi = {10.3389/fmicb.2015.00115},
journal = {Frontiers in Microbiology},
number = ,
volume = 6,
place = {United States},
year = {Wed Feb 11 00:00:00 EST 2015},
month = {Wed Feb 11 00:00:00 EST 2015}
}
Web of Science
Figures / Tables:
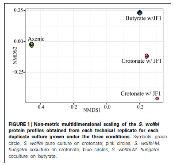 FIGURE 1: Non-metric multidimensional scaling of the S. wolfei protein profiles obtained from each technical replicate for each duplicate culture grown under the three conditions. Symbols: green circle, S. wolfei pure culture on crotonate; pink circles, S. wolfei-M. hungatei coculture on crotonate; blue circles, S. wolfei-M. hungatei coculture on butyrate.
FIGURE 1: Non-metric multidimensional scaling of the S. wolfei protein profiles obtained from each technical replicate for each duplicate culture grown under the three conditions. Symbols: green circle, S. wolfei pure culture on crotonate; pink circles, S. wolfei-M. hungatei coculture on crotonate; blue circles, S. wolfei-M. hungatei coculture on butyrate.
Works referenced in this record:
Flexibility of Syntrophic Enzyme Systems in Desulfovibrio Species Ensures Their Adaptation Capability to Environmental Changes
journal, August 2013
- Meyer, B.; Kuehl, J. V.; Deutschbauer, A. M.
- Journal of Bacteriology, Vol. 195, Issue 21
Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation
journal, December 2006
- Lu, Peng; Vogel, Christine; Wang, Rong
- Nature Biotechnology, Vol. 25, Issue 1
Growth of Syntrophomonas wolfei in pure culture on crotonate
journal, May 1987
- Beaty, P. S.; McInerney, M. J.
- Archives of Microbiology, Vol. 147, Issue 4
Substrate-dependent transcriptomic shifts in Pelotomaculum thermopropionicum grown in syntrophic co-culture with Methanothermobacter thermautotrophicus
journal, September 2009
- Kato, Souichiro; Kosaka, Tomoyuki; Watanabe, Kazuya
- Microbial Biotechnology, Vol. 2, Issue 5
Data extraction from proteomics raw data: An evaluation of nine tandem MS tools using a large Orbitrap data set
journal, September 2012
- Mancuso, Francesco; Bunkenborg, Jakob; Wierer, Michael
- Journal of Proteomics, Vol. 75, Issue 17
Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry
journal, February 2007
- Elias, Joshua E.; Gygi, Steven P.
- Nature Methods, Vol. 4, Issue 3
The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth
journal, April 2007
- McInerney, M. J.; Rohlin, L.; Mouttaki, H.
- Proceedings of the National Academy of Sciences, Vol. 104, Issue 18
Genomic Insights into Syntrophy: The Paradigm for Anaerobic Metabolic Cooperation
journal, October 2012
- Sieber, Jessica R.; McInerney, Michael J.; Gunsalus, Robert P.
- Annual Review of Microbiology, Vol. 66, Issue 1
A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2)
journal, September 2003
- Brekasis, D.
- The EMBO Journal, Vol. 22, Issue 18
Evaluation of Affinity-Tagged Protein Expression Strategies Using Local and Global Isotope Ratio Measurements
journal, July 2009
- Hervey, W. Judson; Khalsa-Moyers, Gurusahai; Lankford, Patricia K.
- Journal of Proteome Research, Vol. 8, Issue 7
Syntrophism among Prokaryotes
book, January 2006
- Schink, Bernhard; Stams, Alfons J. M.
- The Prokaryotes
DTASelect and Contrast: Tools for Assembling and Comparing Protein Identifications from Shotgun Proteomics
journal, February 2002
- Tabb, David L.; McDonald, W. Hayes; Yates, John R.
- Journal of Proteome Research, Vol. 1, Issue 1, p. 21-26
Statistical Analysis of Membrane Proteome Expression Changes in Saccharomyces c erevisiae
journal, September 2006
- Zybailov, Boris; Mosley, Amber L.; Sardiu, Mihaela E.
- Journal of Proteome Research, Vol. 5, Issue 9
Large-scale analysis of the yeast proteome by multidimensional protein identification technology
journal, March 2001
- Washburn, Michael P.; Wolters, Dirk; Yates, John R.
- Nature Biotechnology, Vol. 19, Issue 3
Poly-?-hydroxyalkanoate in Syntrophomonas wolfei
journal, July 1989
- Amos, Dale A.; McInerney, Michael J.
- Archives of Microbiology, Vol. 152, Issue 2
GAF Domains: Two-Billion-Year-Old Molecular Switches that Bind Cyclic Nucleotides
journal, September 2002
- Martinez, S. E.
- Molecular Interventions, Vol. 2, Issue 5
The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production: Syntrophomonas wolfei genome
journal, June 2010
- Sieber, Jessica R.; Sims, David R.; Han, Cliff
- Environmental Microbiology
Variation among Desulfovibrio Species in Electron Transfer Systems Used for Syntrophic Growth
journal, December 2012
- Meyer, B.; Kuehl, J.; Deutschbauer, A. M.
- Journal of Bacteriology, Vol. 195, Issue 5
Experimental Approach for Deep Proteome Measurements from Small-Scale Microbial Biomass Samples
journal, December 2008
- Thompson, Melissa R.; Chourey, Karuna; Froelich, Jennifer M.
- Analytical Chemistry, Vol. 80, Issue 24
An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database
journal, November 1994
- Eng, Jimmy K.; McCormack, Ashley L.; Yates, John R.
- Journal of the American Society for Mass Spectrometry, Vol. 5, Issue 11
Electron transfer in syntrophic communities of anaerobic bacteria and archaea
journal, August 2009
- Stams, Alfons J. M.; Plugge, Caroline M.
- Nature Reviews Microbiology, Vol. 7, Issue 8
Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea
journal, January 2011
- Kaster, A. -K.; Moll, J.; Parey, K.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 7
Coupled Ferredoxin and Crotonyl Coenzyme A (CoA) Reduction with NADH Catalyzed by the Butyryl-CoA Dehydrogenase/Etf Complex from Clostridium kluyveri
journal, November 2007
- Li, F.; Hinderberger, J.; Seedorf, H.
- Journal of Bacteriology, Vol. 190, Issue 3
An Automated Multidimensional Protein Identification Technology for Shotgun Proteomics
journal, December 2001
- Wolters, Dirk A.; Washburn, Michael P.; Yates, John R.
- Analytical Chemistry, Vol. 73, Issue 23
Qscore: An algorithm for evaluating SEQUEST database search results
journal, April 2002
- Moore, Roger E.; Young, Mary K.; Lee, Terry D.
- Journal of the American Society for Mass Spectrometry, Vol. 13, Issue 4
Growth of Synthrophomonas wolfei on unsaturated short chain fatty acids
journal, June 1990
- Amos, DaleA.; McInerney, MichaelJ.
- Archives of Microbiology, Vol. 154, Issue 1
Unusually High Standard Redox Potential of Acrylyl-CoA/ Propionyl-CoA Couple among Enoyl-CoA/Acyl-CoA Couples: A Reason for the Distinct Metabolic Pathway of Propionyl-CoA from Longer Acyl-CoAs
journal, October 1999
- Sato, K.; Nishina, Y.; Setoyama, C.
- Journal of Biochemistry, Vol. 126, Issue 4
Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase
journal, June 2010
- Costa, K. C.; Wong, P. M.; Wang, T.
- Proceedings of the National Academy of Sciences, Vol. 107, Issue 24
RNA-seq reveals cooperative metabolic interactions between two termite-gut spirochete species in co-culture
journal, February 2011
- Rosenthal, Adam Z.; Matson, Eric G.; Eldar, Avigdor
- The ISME Journal, Vol. 5, Issue 7
Physiology, Ecology, Phylogeny, and Genomics of Microorganisms Capable of Syntrophic Metabolism
journal, March 2008
- McInerney, Michael J.; Struchtemeyer, Christopher G.; Sieber, Jessica
- Annals of the New York Academy of Sciences, Vol. 1125, Issue 1
Anaerobic bacterium that degrades fatty acids in syntrophic association with methanogens
journal, August 1979
- McInerney, Michael J.; Bryant, Marvin P.; Pfennig, Norbert
- Archives of Microbiology, Vol. 122, Issue 2
The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism: Importance of interspecies hydrogen and formate transfer
journal, October 2013
- Sieber, Jessica R.; Le, Huynh M.; McInerney, Michael J.
- Environmental Microbiology, Vol. 16, Issue 1
New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere.
journal, January 1976
- Balch, W. E.; Wolfe, R. S.
- Applied and Environmental Microbiology, Vol. 32, Issue 6
Energetics of syntrophic cooperation in methanogenic degradation.
journal, January 1997
- Schink, B.
- Microbiology and molecular biology reviews : MMBR, Vol. 61, Issue 2
PAS Domains: Internal Sensors of Oxygen, Redox Potential, and Light
journal, June 1999
- Taylor, Barry L.; Zhulin, Igor B.
- Microbiology and Molecular Biology Reviews, Vol. 63, Issue 2
Detecting protein and post-translational modifications in single cells with iDentification and qUantification sEparaTion (DUET)
journal, August 2020
- Zhang, Yandong; Sohn, Changho; Lee, Seoyeon
- Communications Biology, Vol. 3, Issue 1
Syntrophism Among Prokaryotes
book, January 2013
- Schink, Bernhard; Stams, Alfons J. M.
- The Prokaryotes
Evidence of reversed electron transport in syntrophic butyrate or benzoate oxidation by Syntrophomonas wolfei and Syntrophus buswellii
journal, July 1994
- Wallrabenstein, Christina; Schink, Bernhard
- Archives of Microbiology, Vol. 162, Issue 1-2
Data extraction from proteomics raw data: An evaluation of nine tandem MS tools using a large Orbitrap data set
journal, September 2012
- Mancuso, Francesco; Bunkenborg, Jakob; Wierer, Michael
- Journal of Proteomics, Vol. 75, Issue 17
An Automated Multidimensional Protein Identification Technology for Shotgun Proteomics
journal, December 2001
- Wolters, Dirk A.; Washburn, Michael P.; Yates, John R.
- Analytical Chemistry, Vol. 73, Issue 23
Experimental Approach for Deep Proteome Measurements from Small-Scale Microbial Biomass Samples
journal, December 2008
- Thompson, Melissa R.; Chourey, Karuna; Froelich, Jennifer M.
- Analytical Chemistry, Vol. 80, Issue 24
Statistical Analysis of Membrane Proteome Expression Changes in Saccharomyces c erevisiae
journal, September 2006
- Zybailov, Boris; Mosley, Amber L.; Sardiu, Mihaela E.
- Journal of Proteome Research, Vol. 5, Issue 9
Large-scale analysis of the yeast proteome by multidimensional protein identification technology
journal, March 2001
- Washburn, Michael P.; Wolters, Dirk; Yates, John R.
- Nature Biotechnology, Vol. 19, Issue 3
RNA-seq reveals cooperative metabolic interactions between two termite-gut spirochete species in co-culture
journal, February 2011
- Rosenthal, Adam Z.; Matson, Eric G.; Eldar, Avigdor
- The ISME Journal, Vol. 5, Issue 7
Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation
journal, December 2006
- Lu, Peng; Vogel, Christine; Wang, Rong
- Nature Biotechnology, Vol. 25, Issue 1
Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry
journal, February 2007
- Elias, Joshua E.; Gygi, Steven P.
- Nature Methods, Vol. 4, Issue 3
Electron transfer in syntrophic communities of anaerobic bacteria and archaea
journal, August 2009
- Stams, Alfons J. M.; Plugge, Caroline M.
- Nature Reviews Microbiology, Vol. 7, Issue 8
Phosphate dysregulation via the XPR1–KIDINS220 protein complex is a therapeutic vulnerability in ovarian cancer
journal, April 2022
- Bondeson, Daniel P.; Paolella, Brenton R.; Asfaw, Adhana
- Nature Cancer, Vol. 3, Issue 6
The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth
journal, April 2007
- McInerney, M. J.; Rohlin, L.; Mouttaki, H.
- Proceedings of the National Academy of Sciences, Vol. 104, Issue 18
Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase
journal, June 2010
- Costa, K. C.; Wong, P. M.; Wang, T.
- Proceedings of the National Academy of Sciences, Vol. 107, Issue 24
Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea
journal, January 2011
- Kaster, A. -K.; Moll, J.; Parey, K.
- Proceedings of the National Academy of Sciences, Vol. 108, Issue 7
A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2)
journal, September 2003
- Brekasis, D.
- The EMBO Journal, Vol. 22, Issue 18
Unusually High Standard Redox Potential of Acrylyl-CoA/ Propionyl-CoA Couple among Enoyl-CoA/Acyl-CoA Couples: A Reason for the Distinct Metabolic Pathway of Propionyl-CoA from Longer Acyl-CoAs
journal, October 1999
- Sato, K.; Nishina, Y.; Setoyama, C.
- Journal of Biochemistry, Vol. 126, Issue 4
The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism: Importance of interspecies hydrogen and formate transfer
journal, October 2013
- Sieber, Jessica R.; Le, Huynh M.; McInerney, Michael J.
- Environmental Microbiology, Vol. 16, Issue 1
The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production: Syntrophomonas wolfei genome
journal, June 2010
- Sieber, Jessica R.; Sims, David R.; Han, Cliff
- Environmental Microbiology
Substrate-dependent transcriptomic shifts in Pelotomaculum thermopropionicum grown in syntrophic co-culture with Methanothermobacter thermautotrophicus
journal, September 2009
- Kato, Souichiro; Kosaka, Tomoyuki; Watanabe, Kazuya
- Microbial Biotechnology, Vol. 2, Issue 5
GAF Domains: Two-Billion-Year-Old Molecular Switches that Bind Cyclic Nucleotides
journal, September 2002
- Martinez, S. E.
- Molecular Interventions, Vol. 2, Issue 5
Syntrophomonas wolfei gen. nov. sp. nov., an Anaerobic, Syntrophic, Fatty Acid-Oxidizing Bacterium
journal, January 1981
- McInerney, M. J.; Bryant, M. P.; Hespell, R. B.
- Applied and Environmental Microbiology, Vol. 41, Issue 4
Classic Spotlight: Electron Bifurcation, a Unifying Concept for Energy Conservation in Anaerobes
journal, April 2016
- Metcalf, William W.
- Journal of Bacteriology, Vol. 198, Issue 9
Regulatory Loop between Redox Sensing of the NADH/NAD+ Ratio by Rex (YdiH) and Oxidation of NADH by NADH Dehydrogenase Ndh in Bacillus subtilis
journal, October 2006
- Gyan, S.; Shiohira, Y.; Sato, I.
- Journal of Bacteriology, Vol. 188, Issue 20
Energetics of syntrophic cooperation in methanogenic degradation
journal, June 1997
- Schink, B.
- Microbiology and Molecular Biology Reviews, Vol. 61, Issue 2
Genomic Insights into Syntrophy: The Paradigm for Anaerobic Metabolic Cooperation
journal, October 2012
- Sieber, Jessica R.; McInerney, Michael J.; Gunsalus, Robert P.
- Annual Review of Microbiology, Vol. 66, Issue 1
Physiology, Ecology, Phylogeny, and Genomics of Microorganisms Capable of Syntrophic Metabolism
journal, March 2008
- McInerney, Michael J.; Struchtemeyer, Christopher G.; Sieber, Jessica
- Annals of the New York Academy of Sciences, Vol. 1125, Issue 1
Works referencing / citing this record:
Label-Free Proteomics of a Defined, Binary Co-culture Reveals Diversity of Competitive Responses Between Members of a Model Soil Microbial System
journal, October 2017
- Chignell, J. F.; Park, S.; Lacerda, C. M. R.
- Microbial Ecology, Vol. 75, Issue 3
Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics
journal, January 2020
- Hao, Liping; Michaelsen, Thomas Yssing; Singleton, Caitlin Margaret
- The ISME Journal, Vol. 14, Issue 4
Methanogenic paraffin degradation proceeds via alkane addition to fumarate by ‘Smithella’ spp. mediated by a syntrophic coupling with hydrogenotrophic methanogens : Methanogenic paraffin-utilizing consortium
journal, June 2016
- Wawrik, Boris; Marks, Christopher R.; Davidova, Irene A.
- Environmental Microbiology, Vol. 18, Issue 8
Comparative proteome analysis of propionate degradation by Syntrophobacter fumaroxidans in pure culture and in coculture with methanogens: Proteome analysis of Syntrophobacter fumaroxidans
journal, April 2018
- Sedano-Núñez, Vicente T.; Boeren, Sjef; Stams, Alfons J. M.
- Environmental Microbiology, Vol. 20, Issue 5
Hydrogen or formate: Alternative key players in methanogenic degradation: Hydrogen/formate as interspecies electron carriers
journal, March 2017
- Schink, Bernhard; Montag, Dominik; Keller, Anja
- Environmental Microbiology Reports, Vol. 9, Issue 3
Elucidating Syntrophic Butyrate-Degrading Populations in Anaerobic Digesters Using Stable-Isotope-Informed Genome-Resolved Metagenomics
journal, August 2019
- Ziels, Ryan M.; Nobu, Masaru K.; Sousa, Diana Z.
- mSystems, Vol. 4, Issue 4
Syntrophic acetate oxidation replaces acetoclastic methanogenesis during thermophilic digestion of biowaste
journal, July 2020
- Dyksma, Stefan; Jansen, Lukas; Gallert, Claudia
- Microbiome, Vol. 8, Issue 1
Membrane Complexes of Syntrophomonas wolfei Involved in Syntrophic Butyrate Degradation and Hydrogen Formation
journal, November 2016
- Crable, Bryan R.; Sieber, Jessica R.; Mao, Xinwei
- Frontiers in Microbiology, Vol. 7
Flavin-Based Electron Bifurcation, Ferredoxin, Flavodoxin, and Anaerobic Respiration With Protons (Ech) or NAD+ (Rnf) as Electron Acceptors: A Historical Review
journal, March 2018
- Buckel, Wolfgang; Thauer, Rudolf K.
- Frontiers in Microbiology, Vol. 9
Concerted Metabolic Shifts Give New Insights Into the Syntrophic Mechanism Between Propionate-Fermenting Pelotomaculum thermopropionicum and Hydrogenotrophic Methanocella conradii
journal, July 2018
- Liu, Pengfei; Lu, Yahai
- Frontiers in Microbiology, Vol. 9
Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics
journal, January 2020
- Hao, Liping; Michaelsen, Thomas Yssing; Singleton, Caitlin Margaret
- The ISME Journal, Vol. 14, Issue 4
Elucidating Syntrophic Butyrate-Degrading Populations in Anaerobic Digesters Using Stable-Isotope-Informed Genome-Resolved Metagenomics
journal, August 2019
- Ziels, Ryan M.; Nobu, Masaru K.; Sousa, Diana Z.
- mSystems, Vol. 4, Issue 4
Comparative Genomics of Syntrophic Branched-Chain Fatty Acid Degrading Bacteria
journal, January 2016
- Narihiro, Takashi; Nobu, Masaru K.; Tamaki, Hideyuki
- Microbes and environments, Vol. 31, Issue 3
Metagenomic Characterization of Candidatus Smithella cisternae Strain M82_1, a Syntrophic Alkane-Degrading Bacteria, Enriched from the Shengli Oil Field
journal, January 2017
- Qin, Qian-Shan; Feng, Ding-Shan; Liu, Peng-Fei
- Microbes and Environments, Vol. 32, Issue 3
Membrane Complexes of Syntrophomonas wolfei Involved in Syntrophic Butyrate Degradation and Hydrogen Formation
journal, November 2016
- Crable, Bryan R.; Sieber, Jessica R.; Mao, Xinwei
- Frontiers in Microbiology, Vol. 7
Flavin-Based Electron Bifurcation, Ferredoxin, Flavodoxin, and Anaerobic Respiration With Protons (Ech) or NAD+ (Rnf) as Electron Acceptors: A Historical Review
journal, March 2018
- Buckel, Wolfgang; Thauer, Rudolf K.
- Frontiers in Microbiology, Vol. 9
Figures / Tables found in this record:

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal