The Ocean Sampling Day Consortium
Abstract
In this study, Ocean Sampling Day was initiated by the EU-funded Micro B3 (Marine Microbial Biodiversity, Bioinformatics, Biotechnology) project to obtain a snapshot of the marine microbial biodiversity and function of the world’s oceans. It is a simultaneous global mega-sequencing campaign aiming to generate the largest standardized microbial data set in a single day. This will be achievable only through the coordinated efforts of an Ocean Sampling Day Consortium, supportive partnerships and networks between sites. This commentary outlines the establishment, function and aims of the Consortium and describes our vision for a sustainable study of marine microbial communities and their embedded functional traits.
- Authors:
- more »
- Publication Date:
- Research Org.:
- Argonne National Laboratory (ANL), Argonne, IL (United States)
- Sponsoring Org.:
- USDOE
- OSTI Identifier:
- 1225000
- Grant/Contract Number:
- AC02-06CH11357
- Resource Type:
- Accepted Manuscript
- Journal Name:
- GigaScience
- Additional Journal Information:
- Journal Volume: 4; Journal Issue: 1; Journal ID: ISSN 2047-217X
- Publisher:
- BioMed Central
- Country of Publication:
- United States
- Language:
- English
- Subject:
- 54 ENVIRONMENTAL SCIENCES; 59 BASIC BIOLOGICAL SCIENCES; Ocean Sampling Day; OSD; biodiversity; genomics; health index; bacteria; microorganism; metagenomics; marine; micro B3; standards
Citation Formats
Kopf, Anna, Bicak, Mesude, Kottmann, Renzo, Schnetzer, Julia, Kostadinov, Ivaylo, Lehmann, Katja, Fernandez-Guerra, Antonio, Jeanthon, Christian, Rahav, Eyal, Ullrich, Matthias, Wichels, Antje, Gerdts, Gunnar, Polymenakou, Paraskevi, Kotoulas, Giorgos, Siam, Rania, Abdallah, Rehab Z., Sonnenschein, Eva C., Cariou, Thierry, O’Gara, Fergal, Jackson, Stephen, Orlic, Sandi, Steinke, Michael, Busch, Julia, Duarte, Bernardo, Caçador, Isabel, Canning-Clode, João, Bobrova, Oleksandra, Marteinsson, Viggo, Reynisson, Eyjolfur, Loureiro, Clara Magalhães, Luna, Gian Marco, Quero, Grazia Marina, Löscher, Carolin R., Kremp, Anke, DeLorenzo, Marie E., Øvreås, Lise, Tolman, Jennifer, LaRoche, Julie, Penna, Antonella, Frischer, Marc, Davis, Timothy, Katherine, Barker, Meyer, Christopher P., Ramos, Sandra, Magalhães, Catarina, Jude-Lemeilleur, Florence, Aguirre-Macedo, Ma Leopoldina, Wang, Shiao, Poulton, Nicole, Jones, Scott, Collin, Rachel, Fuhrman, Jed A., Conan, Pascal, Alonso, Cecilia, Stambler, Noga, Goodwin, Kelly, Yakimov, Michael M., Baltar, Federico, Bodrossy, Levente, Van De Kamp, Jodie, Frampton, Dion M. F., Ostrowski, Martin, Van Ruth, Paul, Malthouse, Paul, Claus, Simon, Deneudt, Klaas, Mortelmans, Jonas, Pitois, Sophie, Wallom, David, Salter, Ian, Costa, Rodrigo, Schroeder, Declan C., Kandil, Mahrous M., Amaral, Valentina, Biancalana, Florencia, Santana, Rafael, Pedrotti, Maria Luiza, Yoshida, Takashi, Ogata, Hiroyuki, Ingleton, Tim, Munnik, Kate, Rodriguez-Ezpeleta, Naiara, Berteaux-Lecellier, Veronique, Wecker, Patricia, Cancio, Ibon, Vaulot, Daniel, Bienhold, Christina, Ghazal, Hassan, Chaouni, Bouchra, Essayeh, Soumya, Ettamimi, Sara, Zaid, El Houcine, Boukhatem, Noureddine, Bouali, Abderrahim, Chahboune, Rajaa, Barrijal, Said, Timinouni, Mohammed, El Otmani, Fatima, Bennani, Mohamed, Mea, Marianna, Todorova, Nadezhda, Karamfilov, Ventzislav, ten Hoopen, Petra, Cochrane, Guy, L’Haridon, Stephane, Bizsel, Kemal Can, Vezzi, Alessandro, Lauro, Federico M., Martin, Patrick, Jensen, Rachelle M., Hinks, Jamie, Gebbels, Susan, Rosselli, Riccardo, De Pascale, Fabio, Schiavon, Riccardo, dos Santos, Antonina, Villar, Emilie, Pesant, Stéphane, Cataletto, Bruno, Malfatti, Francesca, Edirisinghe, Ranjith, Silveira, Jorge A. Herrera, Barbier, Michele, Turk, Valentina, Tinta, Tinkara, Fuller, Wayne J., Salihoglu, Ilkay, Serakinci, Nedime, Ergoren, Mahmut Cerkez, Bresnan, Eileen, Iriberri, Juan, Nyhus, Paul Anders Fronth, Bente, Edvardsen, Karlsen, Hans Erik, Golyshin, Peter N., Gasol, Josep M., Moncheva, Snejana, Dzhembekova, Nina, Johnson, Zackary, Sinigalliano, Christopher David, Gidley, Maribeth Louise, Zingone, Adriana, Danovaro, Roberto, Tsiamis, George, Clark, Melody S., Costa, Ana Cristina, El Bour, Monia, Martins, Ana M., Collins, R. Eric, Ducluzeau, Anne-Lise, Martinez, Jonathan, Costello, Mark J., Amaral-Zettler, Linda A., Gilbert, Jack A., Davies, Neil, Field, Dawn, and Glöckner, Frank Oliver. The Ocean Sampling Day Consortium. United States: N. p., 2015.
Web. doi:10.1186/s13742-015-0066-5.
Kopf, Anna, Bicak, Mesude, Kottmann, Renzo, Schnetzer, Julia, Kostadinov, Ivaylo, Lehmann, Katja, Fernandez-Guerra, Antonio, Jeanthon, Christian, Rahav, Eyal, Ullrich, Matthias, Wichels, Antje, Gerdts, Gunnar, Polymenakou, Paraskevi, Kotoulas, Giorgos, Siam, Rania, Abdallah, Rehab Z., Sonnenschein, Eva C., Cariou, Thierry, O’Gara, Fergal, Jackson, Stephen, Orlic, Sandi, Steinke, Michael, Busch, Julia, Duarte, Bernardo, Caçador, Isabel, Canning-Clode, João, Bobrova, Oleksandra, Marteinsson, Viggo, Reynisson, Eyjolfur, Loureiro, Clara Magalhães, Luna, Gian Marco, Quero, Grazia Marina, Löscher, Carolin R., Kremp, Anke, DeLorenzo, Marie E., Øvreås, Lise, Tolman, Jennifer, LaRoche, Julie, Penna, Antonella, Frischer, Marc, Davis, Timothy, Katherine, Barker, Meyer, Christopher P., Ramos, Sandra, Magalhães, Catarina, Jude-Lemeilleur, Florence, Aguirre-Macedo, Ma Leopoldina, Wang, Shiao, Poulton, Nicole, Jones, Scott, Collin, Rachel, Fuhrman, Jed A., Conan, Pascal, Alonso, Cecilia, Stambler, Noga, Goodwin, Kelly, Yakimov, Michael M., Baltar, Federico, Bodrossy, Levente, Van De Kamp, Jodie, Frampton, Dion M. F., Ostrowski, Martin, Van Ruth, Paul, Malthouse, Paul, Claus, Simon, Deneudt, Klaas, Mortelmans, Jonas, Pitois, Sophie, Wallom, David, Salter, Ian, Costa, Rodrigo, Schroeder, Declan C., Kandil, Mahrous M., Amaral, Valentina, Biancalana, Florencia, Santana, Rafael, Pedrotti, Maria Luiza, Yoshida, Takashi, Ogata, Hiroyuki, Ingleton, Tim, Munnik, Kate, Rodriguez-Ezpeleta, Naiara, Berteaux-Lecellier, Veronique, Wecker, Patricia, Cancio, Ibon, Vaulot, Daniel, Bienhold, Christina, Ghazal, Hassan, Chaouni, Bouchra, Essayeh, Soumya, Ettamimi, Sara, Zaid, El Houcine, Boukhatem, Noureddine, Bouali, Abderrahim, Chahboune, Rajaa, Barrijal, Said, Timinouni, Mohammed, El Otmani, Fatima, Bennani, Mohamed, Mea, Marianna, Todorova, Nadezhda, Karamfilov, Ventzislav, ten Hoopen, Petra, Cochrane, Guy, L’Haridon, Stephane, Bizsel, Kemal Can, Vezzi, Alessandro, Lauro, Federico M., Martin, Patrick, Jensen, Rachelle M., Hinks, Jamie, Gebbels, Susan, Rosselli, Riccardo, De Pascale, Fabio, Schiavon, Riccardo, dos Santos, Antonina, Villar, Emilie, Pesant, Stéphane, Cataletto, Bruno, Malfatti, Francesca, Edirisinghe, Ranjith, Silveira, Jorge A. Herrera, Barbier, Michele, Turk, Valentina, Tinta, Tinkara, Fuller, Wayne J., Salihoglu, Ilkay, Serakinci, Nedime, Ergoren, Mahmut Cerkez, Bresnan, Eileen, Iriberri, Juan, Nyhus, Paul Anders Fronth, Bente, Edvardsen, Karlsen, Hans Erik, Golyshin, Peter N., Gasol, Josep M., Moncheva, Snejana, Dzhembekova, Nina, Johnson, Zackary, Sinigalliano, Christopher David, Gidley, Maribeth Louise, Zingone, Adriana, Danovaro, Roberto, Tsiamis, George, Clark, Melody S., Costa, Ana Cristina, El Bour, Monia, Martins, Ana M., Collins, R. Eric, Ducluzeau, Anne-Lise, Martinez, Jonathan, Costello, Mark J., Amaral-Zettler, Linda A., Gilbert, Jack A., Davies, Neil, Field, Dawn, & Glöckner, Frank Oliver. The Ocean Sampling Day Consortium. United States. https://doi.org/10.1186/s13742-015-0066-5
Kopf, Anna, Bicak, Mesude, Kottmann, Renzo, Schnetzer, Julia, Kostadinov, Ivaylo, Lehmann, Katja, Fernandez-Guerra, Antonio, Jeanthon, Christian, Rahav, Eyal, Ullrich, Matthias, Wichels, Antje, Gerdts, Gunnar, Polymenakou, Paraskevi, Kotoulas, Giorgos, Siam, Rania, Abdallah, Rehab Z., Sonnenschein, Eva C., Cariou, Thierry, O’Gara, Fergal, Jackson, Stephen, Orlic, Sandi, Steinke, Michael, Busch, Julia, Duarte, Bernardo, Caçador, Isabel, Canning-Clode, João, Bobrova, Oleksandra, Marteinsson, Viggo, Reynisson, Eyjolfur, Loureiro, Clara Magalhães, Luna, Gian Marco, Quero, Grazia Marina, Löscher, Carolin R., Kremp, Anke, DeLorenzo, Marie E., Øvreås, Lise, Tolman, Jennifer, LaRoche, Julie, Penna, Antonella, Frischer, Marc, Davis, Timothy, Katherine, Barker, Meyer, Christopher P., Ramos, Sandra, Magalhães, Catarina, Jude-Lemeilleur, Florence, Aguirre-Macedo, Ma Leopoldina, Wang, Shiao, Poulton, Nicole, Jones, Scott, Collin, Rachel, Fuhrman, Jed A., Conan, Pascal, Alonso, Cecilia, Stambler, Noga, Goodwin, Kelly, Yakimov, Michael M., Baltar, Federico, Bodrossy, Levente, Van De Kamp, Jodie, Frampton, Dion M. F., Ostrowski, Martin, Van Ruth, Paul, Malthouse, Paul, Claus, Simon, Deneudt, Klaas, Mortelmans, Jonas, Pitois, Sophie, Wallom, David, Salter, Ian, Costa, Rodrigo, Schroeder, Declan C., Kandil, Mahrous M., Amaral, Valentina, Biancalana, Florencia, Santana, Rafael, Pedrotti, Maria Luiza, Yoshida, Takashi, Ogata, Hiroyuki, Ingleton, Tim, Munnik, Kate, Rodriguez-Ezpeleta, Naiara, Berteaux-Lecellier, Veronique, Wecker, Patricia, Cancio, Ibon, Vaulot, Daniel, Bienhold, Christina, Ghazal, Hassan, Chaouni, Bouchra, Essayeh, Soumya, Ettamimi, Sara, Zaid, El Houcine, Boukhatem, Noureddine, Bouali, Abderrahim, Chahboune, Rajaa, Barrijal, Said, Timinouni, Mohammed, El Otmani, Fatima, Bennani, Mohamed, Mea, Marianna, Todorova, Nadezhda, Karamfilov, Ventzislav, ten Hoopen, Petra, Cochrane, Guy, L’Haridon, Stephane, Bizsel, Kemal Can, Vezzi, Alessandro, Lauro, Federico M., Martin, Patrick, Jensen, Rachelle M., Hinks, Jamie, Gebbels, Susan, Rosselli, Riccardo, De Pascale, Fabio, Schiavon, Riccardo, dos Santos, Antonina, Villar, Emilie, Pesant, Stéphane, Cataletto, Bruno, Malfatti, Francesca, Edirisinghe, Ranjith, Silveira, Jorge A. Herrera, Barbier, Michele, Turk, Valentina, Tinta, Tinkara, Fuller, Wayne J., Salihoglu, Ilkay, Serakinci, Nedime, Ergoren, Mahmut Cerkez, Bresnan, Eileen, Iriberri, Juan, Nyhus, Paul Anders Fronth, Bente, Edvardsen, Karlsen, Hans Erik, Golyshin, Peter N., Gasol, Josep M., Moncheva, Snejana, Dzhembekova, Nina, Johnson, Zackary, Sinigalliano, Christopher David, Gidley, Maribeth Louise, Zingone, Adriana, Danovaro, Roberto, Tsiamis, George, Clark, Melody S., Costa, Ana Cristina, El Bour, Monia, Martins, Ana M., Collins, R. Eric, Ducluzeau, Anne-Lise, Martinez, Jonathan, Costello, Mark J., Amaral-Zettler, Linda A., Gilbert, Jack A., Davies, Neil, Field, Dawn, and Glöckner, Frank Oliver. Fri .
"The Ocean Sampling Day Consortium". United States. https://doi.org/10.1186/s13742-015-0066-5. https://www.osti.gov/servlets/purl/1225000.
@article{osti_1225000,
title = {The Ocean Sampling Day Consortium},
author = {Kopf, Anna and Bicak, Mesude and Kottmann, Renzo and Schnetzer, Julia and Kostadinov, Ivaylo and Lehmann, Katja and Fernandez-Guerra, Antonio and Jeanthon, Christian and Rahav, Eyal and Ullrich, Matthias and Wichels, Antje and Gerdts, Gunnar and Polymenakou, Paraskevi and Kotoulas, Giorgos and Siam, Rania and Abdallah, Rehab Z. and Sonnenschein, Eva C. and Cariou, Thierry and O’Gara, Fergal and Jackson, Stephen and Orlic, Sandi and Steinke, Michael and Busch, Julia and Duarte, Bernardo and Caçador, Isabel and Canning-Clode, João and Bobrova, Oleksandra and Marteinsson, Viggo and Reynisson, Eyjolfur and Loureiro, Clara Magalhães and Luna, Gian Marco and Quero, Grazia Marina and Löscher, Carolin R. and Kremp, Anke and DeLorenzo, Marie E. and Øvreås, Lise and Tolman, Jennifer and LaRoche, Julie and Penna, Antonella and Frischer, Marc and Davis, Timothy and Katherine, Barker and Meyer, Christopher P. and Ramos, Sandra and Magalhães, Catarina and Jude-Lemeilleur, Florence and Aguirre-Macedo, Ma Leopoldina and Wang, Shiao and Poulton, Nicole and Jones, Scott and Collin, Rachel and Fuhrman, Jed A. and Conan, Pascal and Alonso, Cecilia and Stambler, Noga and Goodwin, Kelly and Yakimov, Michael M. and Baltar, Federico and Bodrossy, Levente and Van De Kamp, Jodie and Frampton, Dion M. F. and Ostrowski, Martin and Van Ruth, Paul and Malthouse, Paul and Claus, Simon and Deneudt, Klaas and Mortelmans, Jonas and Pitois, Sophie and Wallom, David and Salter, Ian and Costa, Rodrigo and Schroeder, Declan C. and Kandil, Mahrous M. and Amaral, Valentina and Biancalana, Florencia and Santana, Rafael and Pedrotti, Maria Luiza and Yoshida, Takashi and Ogata, Hiroyuki and Ingleton, Tim and Munnik, Kate and Rodriguez-Ezpeleta, Naiara and Berteaux-Lecellier, Veronique and Wecker, Patricia and Cancio, Ibon and Vaulot, Daniel and Bienhold, Christina and Ghazal, Hassan and Chaouni, Bouchra and Essayeh, Soumya and Ettamimi, Sara and Zaid, El Houcine and Boukhatem, Noureddine and Bouali, Abderrahim and Chahboune, Rajaa and Barrijal, Said and Timinouni, Mohammed and El Otmani, Fatima and Bennani, Mohamed and Mea, Marianna and Todorova, Nadezhda and Karamfilov, Ventzislav and ten Hoopen, Petra and Cochrane, Guy and L’Haridon, Stephane and Bizsel, Kemal Can and Vezzi, Alessandro and Lauro, Federico M. and Martin, Patrick and Jensen, Rachelle M. and Hinks, Jamie and Gebbels, Susan and Rosselli, Riccardo and De Pascale, Fabio and Schiavon, Riccardo and dos Santos, Antonina and Villar, Emilie and Pesant, Stéphane and Cataletto, Bruno and Malfatti, Francesca and Edirisinghe, Ranjith and Silveira, Jorge A. Herrera and Barbier, Michele and Turk, Valentina and Tinta, Tinkara and Fuller, Wayne J. and Salihoglu, Ilkay and Serakinci, Nedime and Ergoren, Mahmut Cerkez and Bresnan, Eileen and Iriberri, Juan and Nyhus, Paul Anders Fronth and Bente, Edvardsen and Karlsen, Hans Erik and Golyshin, Peter N. and Gasol, Josep M. and Moncheva, Snejana and Dzhembekova, Nina and Johnson, Zackary and Sinigalliano, Christopher David and Gidley, Maribeth Louise and Zingone, Adriana and Danovaro, Roberto and Tsiamis, George and Clark, Melody S. and Costa, Ana Cristina and El Bour, Monia and Martins, Ana M. and Collins, R. Eric and Ducluzeau, Anne-Lise and Martinez, Jonathan and Costello, Mark J. and Amaral-Zettler, Linda A. and Gilbert, Jack A. and Davies, Neil and Field, Dawn and Glöckner, Frank Oliver},
abstractNote = {In this study, Ocean Sampling Day was initiated by the EU-funded Micro B3 (Marine Microbial Biodiversity, Bioinformatics, Biotechnology) project to obtain a snapshot of the marine microbial biodiversity and function of the world’s oceans. It is a simultaneous global mega-sequencing campaign aiming to generate the largest standardized microbial data set in a single day. This will be achievable only through the coordinated efforts of an Ocean Sampling Day Consortium, supportive partnerships and networks between sites. This commentary outlines the establishment, function and aims of the Consortium and describes our vision for a sustainable study of marine microbial communities and their embedded functional traits.},
doi = {10.1186/s13742-015-0066-5},
journal = {GigaScience},
number = 1,
volume = 4,
place = {United States},
year = {Fri Jun 19 00:00:00 EDT 2015},
month = {Fri Jun 19 00:00:00 EDT 2015}
}
Web of Science
Figures / Tables:
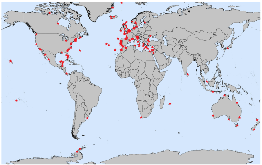 Fig. 1: Map of registered sites for OSD 2014
Fig. 1: Map of registered sites for OSD 2014
Works referenced in this record:
The Earth Microbiome project: successes and aspirations
journal, August 2014
- Gilbert, Jack A.; Jansson, Janet K.; Knight, Rob
- BMC Biology, Vol. 12, Issue 1
A Holistic Approach to Marine Eco-Systems Biology
journal, October 2011
- Karsenti, Eric; Acinas, Silvia G.; Bork, Peer
- PLoS Biology, Vol. 9, Issue 10
An index to assess the health and benefits of the global ocean
journal, August 2012
- Halpern, Benjamin S.; Longo, Catherine; Hardy, Darren
- Nature, Vol. 488, Issue 7413
The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific
journal, March 2007
- Rusch, Douglas B.; Halpern, Aaron L.; Sutton, Granger
- PLoS Biology, Vol. 5, Issue 3
Essential Biodiversity Variables
journal, January 2013
- Pereira, H. M.; Ferrier, S.; Walters, M.
- Science, Vol. 339, Issue 6117
Day-length is central to maintaining consistent seasonal diversity in marine bacterioplankton
journal, May 2010
- Gilbert, Jack; Somerfield, Paul; Temperton, Ben
- Nature Precedings
The founding charter of the Genomic Observatories Network
journal, March 2014
- Davies, Neil; Field, Dawn; Amaral-Zettler, Linda
- GigaScience, Vol. 3, Issue 1
The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific
journal, March 2007
- Rusch, Douglas B.; Halpern, Aaron L.; Sutton, Granger
- PLoS Biology, Vol. 5, Issue 3
A Holistic Approach to Marine Eco-Systems Biology
journal, October 2011
- Karsenti, Eric; Acinas, Silvia G.; Bork, Peer
- PLoS Biology, Vol. 9, Issue 10
Works referencing / citing this record:
Methods for the Study of Marine Biodiversity
book, November 2016
- Costello, Mark J.; Basher, Zeenatul; McLeod, Laura
- The GEO Handbook on Biodiversity Observation Networks
Diversity and Composition of Pelagic Prokaryotic and Protist Communities in a Thin Arctic Sea-Ice Regime
journal, January 2019
- de Sousa, António Gaspar G.; Tomasino, Maria Paola; Duarte, Pedro
- Microbial Ecology, Vol. 78, Issue 2
High diversity and novelty of Actinobacteria isolated from the coastal zone of the geographically remote young volcanic Easter Island, Chile
journal, February 2019
- Sottorff, Ignacio; Wiese, Jutta; Imhoff, Johannes F.
- International Microbiology, Vol. 22, Issue 3
MetaG: a graph-based metagenomic gene analysis for big DNA data
journal, July 2016
- Chowdhury, Linkon; Khan, Mohammad Ibrahim; Deb, Kaushik
- Network Modeling Analysis in Health Informatics and Bioinformatics, Vol. 5, Issue 1
Diversity and oceanic distribution of the Parmales (Bolidophyceae), a picoplanktonic group closely related to diatoms
journal, March 2016
- Ichinomiya, Mutsuo; dos Santos, Adriana Lopes; Gourvil, Priscillia
- The ISME Journal, Vol. 10, Issue 10
Minimum Information about a Biosynthetic Gene cluster
journal, August 2015
- Medema, Marnix H.; Kottmann, Renzo; Yilmaz, Pelin
- Nature Chemical Biology, Vol. 11, Issue 9
An assessment of US microbiome research
journal, January 2016
- Stulberg, Elizabeth; Fravel, Deborah; Proctor, Lita M.
- Nature Microbiology, Vol. 1, Issue 1
The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle
journal, September 2016
- Martínez-Pérez, Clara; Mohr, Wiebke; Löscher, Carolin R.
- Nature Microbiology, Vol. 1, Issue 11
A strong link between marine microbial community composition and function challenges the idea of functional redundancy
journal, June 2018
- Galand, Pierre E.; Pereira, Olivier; Hochart, Corentin
- The ISME Journal, Vol. 12, Issue 10
Metabolic diversity within the globally abundant Marine Group II Euryarchaea offers insight into ecological patterns
journal, January 2019
- Tully, Benjamin J.
- Nature Communications, Vol. 10, Issue 1
Green microalgae in marine coastal waters: The Ocean Sampling Day (OSD) dataset
journal, September 2018
- Tragin, Margot; Vaulot, Daniel
- Scientific Reports, Vol. 8, Issue 1
Diatom diversity through HTS-metabarcoding in coastal European seas
journal, December 2018
- Piredda, Roberta; Claverie, Jean-Michel; Decelle, Johan
- Scientific Reports, Vol. 8, Issue 1
Temporal and spatial dynamics of Bacteria, Archaea and protists in equatorial coastal waters
journal, November 2019
- Chénard, Caroline; Wijaya, Winona; Vaulot, Daniel
- Scientific Reports, Vol. 9, Issue 1
Hidden diversity in the oomycete genus Olpidiopsis is a potential hazard to red algal cultivation and conservation worldwide
journal, November 2019
- Badis, Yacine; Klochkova, Tatyana A.; Brakel, Janina
- European Journal of Phycology, Vol. 55, Issue 2
Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean Long Term Ecological Research site
journal, September 2016
- Piredda, R.; Tomasino, M. P.; D'Erchia, A. M.
- FEMS Microbiology Ecology, Vol. 93, Issue 1
EBI metagenomics in 2016 - an expanding and evolving resource for the analysis and archiving of metagenomic data
journal, November 2015
- Mitchell, Alex; Bucchini, Francois; Cochrane, Guy
- Nucleic Acids Research, Vol. 44, Issue D1
The European Nucleotide Archive in 2019
journal, November 2019
- Amid, Clara; Alako, Blaise T. F.; Balavenkataraman Kadhirvelu, Vishnukumar
- Nucleic Acids Research
Molecular analyses of protists in long-term observation programmes—current status and future perspectives
journal, September 2018
- Stern, Rowena; Kraberg, Alexandra; Bresnan, Eileen
- Journal of Plankton Research, Vol. 40, Issue 5
Censusing marine eukaryotic diversity in the twenty-first century
journal, September 2016
- Leray, Matthieu; Knowlton, Nancy
- Philosophical Transactions of the Royal Society B: Biological Sciences, Vol. 371, Issue 1702
Novel diversity within marine Mamiellophyceae (Chlorophyta) unveiled by metabarcoding
journal, October 2018
- Tragin, Margot; Vaulot, Daniel
- Scientific Reports
Analyses of environmental sequences and two regions of chloroplast genomes revealed the presence of new clades of photosynthetic euglenids in marine environments
journal, October 2019
- Lukešová, Soňa; Karlicki, Michał; Tomečková Hadariová, Lucia
- Environmental Microbiology Reports, Vol. 12, Issue 1
Molecular recognition of the beta‐glucans laminarin and pustulan by a SusD‐like glycan‐binding protein of a marine Bacteroidetes
journal, October 2018
- Mystkowska, Agata Anna; Robb, Craig; Vidal‐Melgosa, Silvia
- The FEBS Journal, Vol. 285, Issue 23
Mantoniella beaufortii and Mantoniella baffinensis sp. nov. (Mamiellales, Mamiellophyceae), two new green algal species from the high arctic 1
journal, November 2019
- Yau, Sheree; Lopes dos Santos, Adriana; Eikrem, Wenche
- Journal of Phycology, Vol. 56, Issue 1
GLOSSary: the GLobal Ocean 16S subunit web accessible resource
journal, November 2018
- Tangherlini, M.; Miralto, M.; Colantuono, C.
- BMC Bioinformatics, Vol. 19, Issue S15
Isolation and genome sequencing of four Arctic marine Psychrobacter strains exhibiting multicopper oxidase activity
journal, February 2016
- Moghadam, Morteza Shojaei; Albersmeier, Andreas; Winkler, Anika
- BMC Genomics, Vol. 17, Issue 1
A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms
journal, November 2018
- Banos, Stefanos; Lentendu, Guillaume; Kopf, Anna
- BMC Microbiology, Vol. 18, Issue 1
BioVeL: a virtual laboratory for data analysis and modelling in biodiversity science and ecology
journal, October 2016
- Hardisty, Alex R.; Bacall, Finn; Beard, Niall
- BMC Ecology, Vol. 16, Issue 1
The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation
journal, September 2016
- Buttigieg, Pier Luigi; Pafilis, Evangelos; Lewis, Suzanna E.
- Journal of Biomedical Semantics, Vol. 7, Issue 1
Performance evaluation of a new custom, multi-component DNA isolation method optimized for use in shotgun metagenomic sequencing-based aerosol microbiome research
journal, January 2020
- Bøifot, Kari Oline; Gohli, Jostein; Moen, Line Victoria
- Environmental Microbiome, Vol. 15, Issue 1
The Genomic Observatories Metadatabase (GeOMe): A new repository for field and sampling event metadata associated with genetic samples
journal, August 2017
- Deck, John; Gaither, Michelle R.; Ewing, Rodney
- PLOS Biology, Vol. 15, Issue 8
The importance of standardization for biodiversity comparisons: A case study using autonomous reef monitoring structures (ARMS) and metabarcoding to measure cryptic diversity on Mo’orea coral reefs, French Polynesia
journal, April 2017
- Ransome, Emma; Geller, Jonathan B.; Timmers, Molly
- PLOS ONE, Vol. 12, Issue 4
Registry of samples and environmental context from the Ocean Sampling Day 2014
dataset, January 2015
- Ocean Sampling Day Consortium, Participants
- PANGAEA, Data Publisher for Earth & Environmental Science, 7078 data points
Implementing and Innovating Marine Monitoring Approaches for Assessing Marine Environmental Status
journal, November 2016
- Danovaro, Roberto; Carugati, Laura; Berzano, Marco
- Frontiers in Marine Science, Vol. 3
DNA Sequencing as a Tool to Monitor Marine Ecological Status
journal, May 2017
- Goodwin, Kelly D.; Thompson, Luke R.; Duarte, Bernardo
- Frontiers in Marine Science, Vol. 4
A Conceptual Framework for Developing the Next Generation of Marine OBservatories (MOBs) for Science and Society
journal, September 2018
- Crise, Alessandro; Ribera d’Alcalà, Maurizio; Mariani, Patrizio
- Frontiers in Marine Science, Vol. 5
Bolidophyceae, a Sister Picoplanktonic Group of Diatoms – A Review
journal, October 2018
- Kuwata, Akira; Yamada, Kazumasa; Ichinomiya, Mutsuo
- Frontiers in Marine Science, Vol. 5
Global Observational Needs and Resources for Marine Biodiversity
journal, July 2019
- Canonico, Gabrielle; Buttigieg, Pier Luigi; Montes, Enrique
- Frontiers in Marine Science, Vol. 6
A Response to Scientific and Societal Needs for Marine Biological Observations
journal, July 2019
- Bax, Nicholas J.; Miloslavich, Patricia; Muller-Karger, Frank Edgar
- Frontiers in Marine Science, Vol. 6
Access to Marine Genetic Resources (MGR): Raising Awareness of Best-Practice Through a New Agreement for Biodiversity Beyond National Jurisdiction (BBNJ)
journal, September 2019
- Rabone, Muriel; Harden-Davies, Harriet; Collins, Jane Eva
- Frontiers in Marine Science, Vol. 6
Expanding the World of Marine Bacterial and Archaeal Clades
journal, January 2016
- Yilmaz, Pelin; Yarza, Pablo; Rapp, Josephine Z.
- Frontiers in Microbiology, Vol. 6
Novel Widespread Marine Oomycetes Parasitising Diatoms, Including the Toxic Genus Pseudo-nitzschia: Genetic, Morphological, and Ecological Characterisation
journal, December 2018
- Garvetto, Andrea; Nézan, Elisabeth; Badis, Yacine
- Frontiers in Microbiology, Vol. 9
Citizen Bio-Optical Observations from Coast- and Ocean and Their Compatibility with Ocean Colour Satellite Measurements
journal, October 2016
- Busch, Julia; Bardaji, Raul; Ceccaroni, Luigi
- Remote Sensing, Vol. 8, Issue 11
Global distribution and diversity of Chaetoceros (Bacillariophyta, Mediophyceae): integration of classical and novel strategies
journal, January 2019
- De Luca, Daniele; Kooistra, Wiebe H. C. F.; Sarno, Diana
- PeerJ, Vol. 7
Novel diversity within marine Mamiellophyceae (Chlorophyta) unveiled by metabarcoding
journal, March 2019
- Tragin, Margot; Vaulot, Daniel
- Scientific Reports, Vol. 9, Issue 1
Molecular recognition of the beta-glucans laminarin and pustulan by a SusD-like glycan-binding protein of a marine Bacteroidetes
text, January 2018
- Mystkowska, Agata Anna; Robb, Craig; Vidal-Melgosa, Silvia
- Deutsches Elektronen-Synchrotron, DESY, Hamburg
Gene Expression Changes and Community Turnover Differentially Shape the Global Ocean Metatranscriptome
journal, November 2019
- Salazar, Guillem; Paoli, Lucas; Alberti, Adriana
- Cell, Vol. 179, Issue 5
Intracellular Infection of Diverse Diatoms by an Evolutionary Distinct Relative of the Fungi
journal, December 2019
- Chambouvet, Aurélie; Monier, Adam; Maguire, Finlay
- Current Biology, Vol. 29, Issue 23
Diversity and oceanic distribution of the Parmales (Bolidophyceae), a picoplanktonic group closely related to diatoms
journal, March 2016
- Ichinomiya, Mutsuo; dos Santos, Adriana Lopes; Gourvil, Priscillia
- The ISME Journal, Vol. 10, Issue 10
Diatom diversity through HTS-metabarcoding in coastal European seas
journal, December 2018
- Piredda, Roberta; Claverie, Jean-Michel; Decelle, Johan
- Scientific Reports, Vol. 8, Issue 1
Temporal and spatial dynamics of Bacteria, Archaea and protists in equatorial coastal waters
journal, November 2019
- Chénard, Caroline; Wijaya, Winona; Vaulot, Daniel
- Scientific Reports, Vol. 9, Issue 1
EBI metagenomics in 2016 - an expanding and evolving resource for the analysis and archiving of metagenomic data
journal, November 2015
- Mitchell, Alex; Bucchini, Francois; Cochrane, Guy
- Nucleic Acids Research, Vol. 44, Issue D1
Censusing marine eukaryotic diversity in the twenty-first century
journal, September 2016
- Leray, Matthieu; Knowlton, Nancy
- Philosophical Transactions of the Royal Society B: Biological Sciences, Vol. 371, Issue 1702
Novel diversity within marine Mamiellophyceae (Chlorophyta) unveiled by metabarcoding
journal, October 2018
- Tragin, Margot; Vaulot, Daniel
- Scientific Reports
Visualizing the invisible: class excursions to ignite children’s enthusiasm for microbes
journal, May 2020
- McGenity, Terry J.; Gessesse, Amare; Hallsworth, John E.
- Microbial Biotechnology, Vol. 13, Issue 4
MyOSD 2014: Evaluating Oceanographic Measurements Contributed by Citizen Scientists in Support of Ocean Sampling Day
journal, March 2016
- Schnetzer, Julia; Kopf, Anna; Bietz, Matthew J.
- Journal of Microbiology & Biology Education, Vol. 17, Issue 1
GLOSSary: the GLobal Ocean 16S subunit web accessible resource
journal, November 2018
- Tangherlini, M.; Miralto, M.; Colantuono, C.
- BMC Bioinformatics, Vol. 19, Issue S15
A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms
journal, November 2018
- Banos, Stefanos; Lentendu, Guillaume; Kopf, Anna
- BMC Microbiology, Vol. 18, Issue 1
BioVeL: a virtual laboratory for data analysis and modelling in biodiversity science and ecology
journal, October 2016
- Hardisty, Alex R.; Bacall, Finn; Beard, Niall
- BMC Ecology, Vol. 16, Issue 1
The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation
journal, September 2016
- Buttigieg, Pier Luigi; Pafilis, Evangelos; Lewis, Suzanna E.
- Journal of Biomedical Semantics, Vol. 7, Issue 1
ELIXIR pilot action: Marine metagenomics – towards a domain specific set of sustainable services
journal, January 2017
- Robertsen, Espen Mikal; Denise, Hubert; Mitchell, Alex
- F1000Research, Vol. 6
Assessment of Common and Emerging Bioinformatics Pipelines for Targeted Metagenomics
journal, January 2017
- Siegwald, Léa; Touzet, Hélène; Lemoine, Yves
- PLOS ONE, Vol. 12, Issue 1
Overview of Integrative Assessment of Marine Systems: The Ecosystem Approach in Practice
journal, March 2016
- Borja, Angel; Elliott, Michael; Andersen, Jesper H.
- Frontiers in Marine Science, Vol. 3
Assessing Future Flood Hazards for Adaptation Planning in a Northern European Coastal Community
journal, May 2016
- Sorensen, Carlo; Broge, Niels H.; Molgaard, Mads R.
- Frontiers in Marine Science, Vol. 3
Implementing and Innovating Marine Monitoring Approaches for Assessing Marine Environmental Status
journal, November 2016
- Danovaro, Roberto; Carugati, Laura; Berzano, Marco
- Frontiers in Marine Science, Vol. 3
DNA Sequencing as a Tool to Monitor Marine Ecological Status
journal, May 2017
- Goodwin, Kelly D.; Thompson, Luke R.; Duarte, Bernardo
- Frontiers in Marine Science, Vol. 4
Novel Widespread Marine Oomycetes Parasitising Diatoms, Including the Toxic Genus Pseudo-nitzschia: Genetic, Morphological, and Ecological Characterisation
journal, December 2018
- Garvetto, Andrea; Nézan, Elisabeth; Badis, Yacine
- Frontiers in Microbiology, Vol. 9
Understanding Marine Microbes, the Driving Engines of the Ocean
journal, January 2016
- Kopf, Anna; Schnetzer, Julia; Glöckner, Frank Oliver
- Frontiers for Young Minds, Vol. 4
Surfing and dining on the “plastisphere”: Microbial life on plastic marine debris
journal, December 2017
- Quero, Grazia Marina; Luna, Gian Marco
- Advances in Oceanography and Limnology, Vol. 8, Issue 2

 Search WorldCat to find libraries that may hold this journal
Search WorldCat to find libraries that may hold this journal