Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models
- Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States)
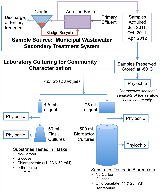
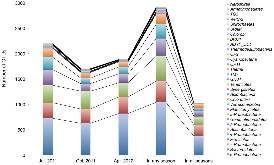
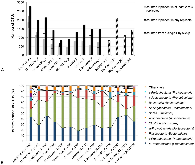
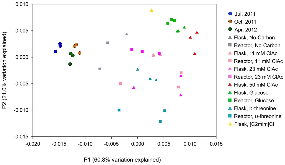
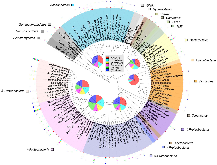
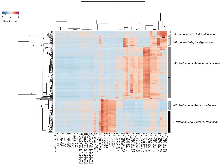
We are developing a laboratory-scale model to improve our understanding and capacity to assess the biological risks of genetically engineered bacteria and their genetic elements in the natural environment. Our hypothetical scenario concerns an industrial bioreactor failure resulting in the introduction of genetically engineered bacteria to a downstream municipal wastewater treatment plant (MWWTP). As the first step towards developing a model for this scenario, we sampled microbial communities from the aeration basin of a MWWTP at three seasonal time points. Having established a baseline for community composition, we investigated how the community changed when propagated in the laboratory, including cell culture media conditions that could provide selective pressure in future studies. Specifically, using PhyloChip 16S-rRNA-gene targeting microarrays, we compared the compositions of sampled communities to those of inocula propagated in the laboratory in simulated wastewater conditionally amended with various carbon sources (glucose, chloroacetate, D-threonine) or the ionic liquid 1-ethyl-3-methylimidazolium chloride ([C2mim]Cl). Proteobacteria, Bacteroidetes, and Actinobacteria were predominant in both aeration basin and laboratory-cultured communities. Laboratory-cultured communities were enriched in γ-Proteobacteria. Enterobacteriaceae, and Aeromonadaceae were enriched by glucose, Pseudomonadaceae by chloroacetate and D-threonine, and Burkholderiacea by high (50 mM) concentrations of chloroacetate. Microbial communities cultured with chloroacetate and D-threonine were more similar to sampled field communities than those cultured with glucose or [C2mim]Cl. Although observed relative richness in operational taxonomic units (OTUs) was lower for laboratory cultures than for field communities, both flask and reactor systems supported phylogenetically diverse communities. These results importantly provide a foundation for laboratory models of industrial bioreactor failure scenarios.
- Research Organization:
- Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States)
- Sponsoring Organization:
- USDOE National Nuclear Security Administration (NNSA), Office of Defense Nuclear Nonproliferation
- Grant/Contract Number:
- AC02-05CH11231
- OSTI ID:
- 1511406
- Journal Information:
- PLoS ONE, Vol. 9, Issue 8; ISSN 1932-6203
- Publisher:
- Public Library of ScienceCopyright Statement
- Country of Publication:
- United States
- Language:
- English
Web of Science
Effect of hydraulic retention time on microbial community structure in wastewater treatment electro-bioreactors
|
journal | March 2018 |
Comparative diversity of microbiomes and Resistomes in beef feedlots, downstream environments and urban sewage influent
|
journal | August 2019 |
Metagenomic analysis of an ecological wastewater treatment plant’s microbial communities and their potential to metabolize pharmaceuticals
|
journal | July 2016 |
Similar Records
Comparison Of A Laboratory Consortium That Dechlorinates TCE To Ethene To The Field Community From Which It Was Derived
A Dechlorinating Community Resulting From In Situ Biostimulation of a TCE-contaminated Deep Fracture Basalt Aquifer