In Silico Identification of Microbial Partners to Form Consortia with Anaerobic Fungi
- Univ. of California, Santa Barbara, CA (United States). Department of Chemical Engineering
- Univ. of California, Santa Barbara, CA (United States). Department of Computer Science
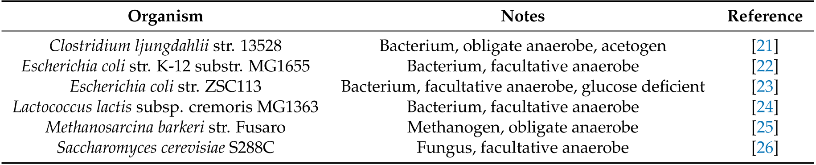
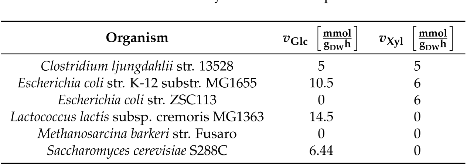
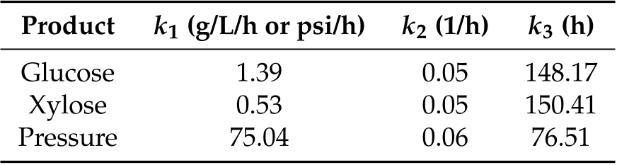
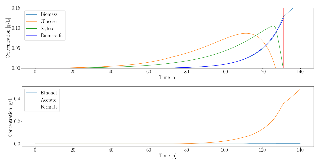
Lignocellulose is an abundant and renewable resource that holds great promise for sustainable bioprocessing. However, unpretreated lignocellulose is recalcitrant to direct utilization by most microbes. Current methods to overcome this barrier include expensive pretreatment steps to liberate cellulose and hemicellulose from lignin. Anaerobic gut fungi possess complex cellulolytic machinery specifically evolved to decompose crude lignocellulose, but they are not yet genetically tractable and have not been employed in industrial bioprocesses. Here, we aim to exploit the biomass-degrading abilities of anaerobic fungi by pairing them with another organism that can convert the fermentable sugars generated from hydrolysis into bioproducts. By combining experiments measuring the amount of excess fermentable sugars released by the fungal enzymes acting on crude lignocellulose, and a novel dynamic flux balance analysis algorithm, we screened potential consortia partners by qualitative suitability. Microbial growth simulations reveal that the fungus Anaeromyces robustus is most suited to pair with either the bacterium Clostridia ljungdahlii or the methanogen Methanosarcina barkeri—both organisms also found in the rumen microbiome. By capitalizing on simulations to screen six alternative organisms, valuable experimental time is saved towards identifying stable consortium members. This approach is also readily generalizable to larger systems and allows one to rationally select partner microbes for formation of stable consortia with non-model microbes like anaerobic fungi.
- Research Organization:
- Univ. of California, Santa Barbara, CA (United States)
- Sponsoring Organization:
- USDOE Office of Science (SC), Biological and Environmental Research (BER). Biological Systems Science Division
- Grant/Contract Number:
- SC0010352
- OSTI ID:
- 1485152
- Journal Information:
- Processes, Vol. 6, Issue 1; ISSN 2227-9717
- Publisher:
- Multidisciplinary Digital Publishing Institute (MDPI)Copyright Statement
- Country of Publication:
- United States
- Language:
- English
Web of Science
Biomass-degrading enzymes are catabolite repressed in anaerobic gut fungi
|
journal | October 2018 |
Special Issue: Microbial Community Modeling: Prediction of Microbial Interactions and Community Dynamics
|
journal | April 2018 |
Predicting the Longitudinally and Radially Varying Gut Microbiota Composition Using Multi-Scale Microbial Metabolic Modeling
|
journal | June 2019 |
Similar Records
Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose
Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose