HipMCL: a high-performance parallel implementation of the Markov clustering algorithm for large-scale networks
- Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States). Computational Research Division
- USDOE Joint Genome Institute (JGI), Walnut Creek, CA (United States)
- Centre for Research & Technology Hellas, Thessalonica (Greece). Biological Computation & Process Lab. Chemical Process & Energy Resources Inst.
- Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States). Computational Research Division; Univ. of California, Berkeley, CA (United States). Dept. of Electrical Engineering and Computer Sciences
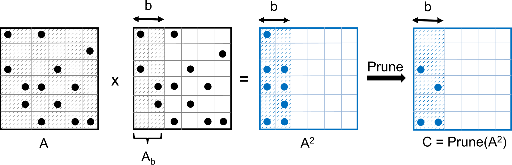
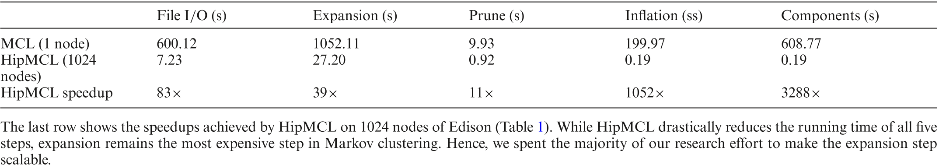
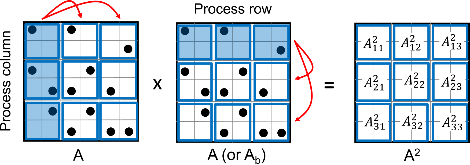
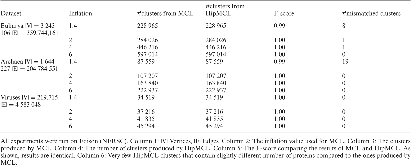
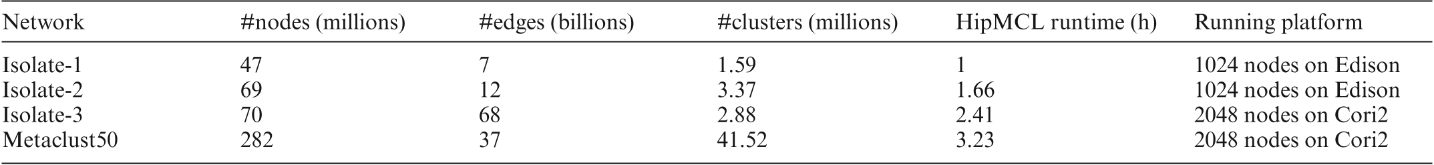
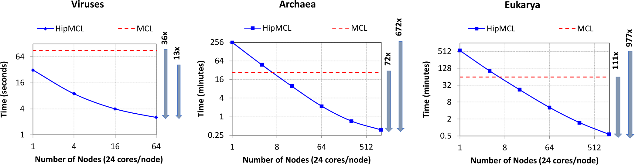
Biological networks capture structural or functional properties of relevant entities such as molecules, proteins or genes. Characteristic examples are gene expression networks or protein–protein interaction networks, which hold information about functional affinities or structural similarities. Such networks have been expanding in size due to increasing scale and abundance of biological data. While various clustering algorithms have been proposed to find highly connected regions, Markov Clustering (MCL) has been one of the most successful approaches to cluster sequence similarity or expression networks. Despite its popularity, MCL’s scalability to cluster large datasets still remains a bottleneck due to high running times and memory demands. In this paper, we present High-performance MCL (HipMCL), a parallel implementation of the original MCL algorithm that can run on distributed-memory computers. We show that HipMCL can efficiently utilize 2000 compute nodes and cluster a network of ~70 million nodes with ~68 billion edges in ~2.4 h. By exploiting distributed-memory environments, HipMCL clusters large-scale networks several orders of magnitude faster than MCL and enables clustering of even bigger networks. Finally, HipMCL is based on MPI and OpenMP and is freely available under a modified BSD license.
- Research Organization:
- Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA (United States)
- Sponsoring Organization:
- USDOE Office of Science (SC), Advanced Scientific Computing Research (ASCR); USDOE National Nuclear Security Administration (NNSA)
- Grant/Contract Number:
- AC02-05CH11231
- OSTI ID:
- 1439241
- Journal Information:
- Nucleic Acids Research, Vol. 46, Issue 6; ISSN 0305-1048
- Publisher:
- Oxford University PressCopyright Statement
- Country of Publication:
- United States
- Language:
- English
Web of Science
Similar Records
Multi-node and Multi-core Performance Studies of a Monte Carlo code RMC
Roofline Analysis in the Intel® Advisor to Deliver Optimized Performance for applications on Intel® Xeon Phi™ Processor